
Purification and enzymatic properties of arsenic resistance protein ArsH from heterogeneous expression in E.coli BL21
WU Xue-ling(吴学玲)1, 2, MIAO Bo(苗 博) 1, 2, HAN Jian(韩 剑) 1, 2, HU Qi(胡 琪) 1, 2,
ZENG Jia(曾 嘉) 1, 2, LIU Yuan-dong(刘元东) 1, 2, QIU Guan-zhou(邱冠周)1, 2
1. School of Resources Processing and Bioengineering, Central South University, Changsha 410083, China;
2. Key Laboratory of Biometallurgy of Ministry of Education, Central South University, Changsha 410083, China
Received 8 September 2009; accepted 22 March 2010
Abstract: Four arsenic-resistance genes (arsB, arsC, arsH, arsR) have been discovered in Acidithiobacillus ferrooxidans. Their gene sequences have been identified and three different arsenic-resistance mechanisms have been elucidated. However, the function of the arsH gene in At. ferrooxidans remains unclear. In order to evaluate the function of the arsH gene, we cloned it and expressed it in Escherichia coli. The protein was purified and its relative molecular mass was determined by SDS-PAGE (Sodium dodecyl sulfate-polyacrylamide gel electrophoresis). The results indicated that the relative molecular mass of the purified ArsH was approximately 29 kDa. The purified protein ArsH from E.coli BL21 was a flavoprotein that oxidized in vitro NADPH with an optimal pH of 6.4.
Key words: Acidithiobacillus ferrooxidans; protein ArsH; expression and purification; FMN reductase; NADPH
1 Introduction
At. ferrooxidans, an acidophilic chemolithotrophic bacterium (the optimal pH for growth of 1.8-2.2), utilizes the oxidation of ferrous iron to ferric iron, or of sulfur (and reduced sulfur compounds) to sulfuric acid, as its energy source. At. ferrooxidans has important applications in biohydrometallurgy. During the bio-oxidation of sulfides, a large quantity of arsenic is released into the surrounding solution and this can be highly bactericidal. At. ferrooxidans possesses enzymes that can oxidize As(III) to As(V) or reduce As(V) to As(III). Arsenic resistance genes are widespread[1]. Arsenite detoxification mechanisms are nearly ubiquitous in microorganisms and are often similar. The first step is the reduction of arsenate to arsenite[2], followed by arsenic removal from the cytosol[3]. At present, an operon containing four arsenic-resistance genes (arsB, arsC, arsH, arsR) has been discovered in At. ferrooxidans. Genes for arsenic resistance are located on the chromosomes of At. ferrooxidans and have been cloned and sequenced[3-5]. Three of them (arsB, arsC, and arsR) have been studied and their arsenic-resistance mechanisms have been elucidated[6-11]. ArsB is an inner membrane channel protein, which participates in the pumping of arsenite across the inner membrane[4, 6-9, 12]. ArsC is an enzyme that reduces less toxic arsenate [As(V)] to the more toxic arsenite [As(III)][2], then arsenite can be pumped out of the cell through the action of ArsB. The activity of ArsC is closely coupled with efflux of arsenite from the cells[7] so that intracellular accumulation of toxic arsenite is prevented[3, 10]. The ArsR protein is a trans-acting repressor protein that acts as a negative regulator of the ars operon[7, 12] that controls the ars operon expression. Another gene, arsH, was first found in Yersinia enterocolitica in 1996[13]. The presence of the arsH gene was essential for resistance to arsenite and arsenate in some microorganisms such as Y. enterocolitica[13], but the arsH gene from At. ferrooxidans showed no arsenic resistance phenotype under the condition tested by using E.coli cells carrying arsH gene from At. ferrooxidans ATCC 23270[4]. Because the ΔpH of At.ferrooxidans between the cytoplasm and the exterior is different from that of E. coli[14], the role of the arsH gene in arsenic resistance remains uncertain in At. ferrooxidans cells. Although the specific biological function of the arsH gene is unclear, the crystal structure of an ArsH protein was presented and NADPH- dependent FMN reductase activity was assayed from Shigella flexneri[15].
In this work, the arsH gene from At. ferrooxidans DX5 was cloned and successfully expressed in E. coli. The ArsH protein was purified by one-step affinity chromatography. The enzymatic activity was analyzed and the optimal pH for FMN reductase activity of the purified ArsH from E.coli was demonstrated.
2 Experimental
2.1 Strains and reagents
At. ferrooxidans DX5 was isolated from an acid mineral drainage sample collected from Dexing Copper Mine, Jiangxi Province, China, which was deposited in the China Center for Type Culture Collection (CCTCC) (Accession No. CCTCC CSU 208059). Top10 competent cells, E. coli strain BL21 (DE3) pLysS, were purchased from Invitrogen (San Diego, USA). Taq DNA polymerase, restriction enzymes and T4 DNA ligase were purchased from MBI Fermentas (Germany). NADPH was purchased from Sigma (St. Louis, USA). Primers were synthesized by Sangon Biotechnology Corp. (Shanghai, China). All other chemicals were of analytical grade and obtained from standard commercial resources.
2.2 Cloning arsH gene from At. ferrooxidans DX5
Genomic DNA was extracted from At. ferrooxidans DX5 which was grown for 72 h at 30 °C in 9K liquid medium with the initial pH adjusted to 1.9-2.1 using 1?1 H2SO4[16]. The PCR (polymerase chain reaction) primer sequences (F: 5'-CGCGCGAATTCAGGAGGAATTT AAAATGAGAGGATCGCATCACCATCACCATCACGTCATCATGTCTGGAAATTTGCCCAATACC-3',and R: 5'-CTGCAGGTCGACTTATAGGTTCTGTAG ATTGACGCGCCGGGAAAG-3') were designed to add six continuous histidine codons to the 5' end of the amplicon and PCR was performed using a 25 mL PCR reaction system and conditions comprising a denaturation step of 60 s at 94 °C, 30 cycles of denaturation for 45 s at 94 °C, annealing for 40 s at 54 °C, and elongation for 90 s at 72 °C. The amplified PCR products were gel purified, double digested with BamH I and EcoR I and ligated into a pLM1 expression vector resulting in the pLM1::arsH plasmid[17]. The identified positive colony was inoculated in Luria Bertani (LB) containing 50 mg/L ampicillin, and the resultant plasmid pLM1::arsH was isolated from harvested bacteria cells using a plasmid extraction kit. The procedures for preparation of plasmid, digestion with restriction enzymes, ligation and transformation all followed the protocol provided by the manufacturer. The recombinant plasmids were then transformed into E. coli strain BL21(DE3) pLysS competent cells for expression.
2.3 Expression and purification of ArsH protein from
E.coli BL21
The procedures for the protein expression were essentially the same as that previously described[18-19]. Briefly, the strain with the recombinant plasmids was inoculated into 500 mL of LB medium containing 100 μg/mL ampicillin and grown overnight at 37 °C. When an optical density OD600 reached 0.6, expression of the protein was induced by the addition of isopropyl-β- D-thiogalactoside (IPTG) to a final concentration of 0.5 mmol/L. The cells were harvested by centrifugation and the cell pellet was washed with an equal volume of distilled water. The cells were again collected by centrifugation, suspended in start buffer containing 20 mmol/L potassium phosphate, (pH 7.4) and 0.5 mol/L NaCl, incubated with 5 mg lysozyme at room temperature for 0.5 h. The harvested cells were lysed by sonication. Insoluble materials (including bodies and other cell debris) were removed by centrifugation. The ArsH protein was purified using HisTrap chelating HP column (GE Healthcare Ltd) which had been pre-charged with Ni2+ ions. The HisTrap column was first washed by five column volumes of distilled water to avoid air bubbles, and then by five column volumes of start buffer to equilibrate the column. The pretreated sample was applied to the HisTrap column after filtering it through 0.22 μm filter. The column was washed with five column volumes of start buffer followed with five column volumes of wash buffer containing 20 mmol/L potassium phosphate, (pH 7.4), 0.5 mol/L NaCl, 50 mmol/L imidazole. Subsequently, the recombinant proteins were eluted with the same buffer containing 500 mmol/L imidazole. The purified protein was stored in a freezer at -80 °C.
2.4 Protein concentration
The protein concentrations were determined by Bradford method using bovine serum albumin as the concentration standard.
2.5 Identification and activity assay of ArsH protein
from E.coli BL21
SDS-PAGE of the protein samples was carried out according to the method of LAEMMLI[20] to determine the relative molecular mass of the ArsH protein. The gels were stained using Coomassie brilliant blue R-250 and distained by washing with 10% acetic acid. ArsH was measured for NADPH?FMN oxidoreductase activity in a reaction mixture (600 μL) consisting of 0.25 mmol/L NADPH, 50 mmol/L KCl, 1 mmol/L EDTA, sodium hydrogen phosphate-citric acid buffer solution and 10 nmol/L ArsH protein at room temperature (30 °C). The pH values of the culture medium (pH 2.2–8.0 with 0.2 pH intervals) were tested for optimal activity of the recombinant proteins. The reactions were run in different buffers for 10 min at room temperature. NADPH oxidation was determined by monitoring the decrease in absorbance at 340 nm. Reactions were initiated by the addition of ArsH protein and control reaction was performed in the absence of the protein. All spectra were recorded with a Techcomp UV-2300 spectrophotometer at room temperature. All assays were repeated three times and arsenite or arsenate did not participate in the reaction.
2.6 UV-visible scanning of ArsH protein from E.coli
BL21
UV-vis absorption was recorded with a Techcomp UV-2300 spectrophotometer at room temperature. A sample of the purified ArsH (10 ?mol/L) was prepared in 20 mmol/L phosphate buffer containing 0.5 mol/L NaCl at pH 7.4. Cysteine (0.5 mmol/L) was added and incubated for 30 min. To adjust the protein concentration, the same volume of buffer was added to the reference cell.
2.7 Molecular structure modeling of ArsH protein
from At. ferrooxidans DX5
The amino acid sequences of the ArsH protein were deduced from the arsH gene sequences which were cloned from the At. ferrooxidans DX5 genomic DNA. All simulations were performed on the Dell Precision 470 workstation with Redhat Linux system using InsightII software package developed by Accelrys Software Inc. The homology module was used to build the initial 3D model of the ArsH protein from At. ferrooxidans DX5. The first step was to search for a number of related sequences to find a related protein as a template by the FASTA program. Then, the program Modeler was performed to build the 3D structure of this protein. Modeler is an implementation of an automated approach to comparative modeling by satisfaction of spatial restraints. Through the procedure, an initial model was completed.
The initial model was further improved by energy minimization (EM). After performing 600 steps of conjugate gradient (CG) minimization, molecular dynamics (MD) simulation was carried out to examine the quality of the model structures, by checking their stability via performing 120 ps simulations at a constant temperature of 298 K. An explicit solvent model water
was used, and the homology solvent model was constructed with a 20 ? water cap from the center of mass of this protein. Finally, a conjugate gradient energy minimization of full protein was performed until the root mean square (RMS) gradient energy was lower than 4.2 J/(mol??). All simulations mentioned above were accomplished by using Discover_3 software module. In this step, the quality of the initial model was improved.
After the optimization procedure, the structure was assessed by ProStat. The ProStat module of Insight II identifies and lists the number of instances where structural features differ significantly from the average values calculated from known proteins. This is especially useful in the final phase of the protein structure modeling.
3 Results
3.1 Cloning of arsH gene from At. ferrooxidans DX5
The arsH gene was amplified from the genomic DNA of At. ferrooxidans DX5 using the primers designed on the basis of the nucleotide sequences of the arsH gene from At. ferrooxidans ATCC 23270. Expression plasmid of pLM11::arsH was constructed, and their sequences were verified by DNA sequencing. The plasmids were then transformed into E. coli BL21 (DE3) pLysS for expression.
3.2 Expression and purification of ArsH protein from
E.coli BL21
The plasmids were transformed into BL21 (DE3) pLysS and the expression of the ArsH protein from E.coli BL21 was induced with IPTG. The N-terminal 6x His-tagged ArsH proteins expressed at a high level. The ArsH protein was observed to be a yellow protein, indicating that the flavin was still bound to the purified protein. The purities of the purified ArsH were further examined by SDS-PAGE and single bands corresponding to a 29 kDa protein were observed with 94% purity (Fig.1).
3.3 Identification and UV-visible scanning of ArsH
protein from E.coli BL21
The purified ArsH showed a yellow coloration and was stable in a mixture of glycerol at 0 °C. The molecular size of the ArsH protein was about 29 kDa as observed by SDS-PAGE. The UV-visible spectrum of the ArsH protein from E.coli BL21 is shown in Fig.2. The result showed that it had two absorption peaks at 448 nm and 382 nm, respectively, which were characteristics for proteins containing FMN. The result was in agreement with the previously reported spectra for native flavoprotein from Entamoeba histolytica[21] and Sinorhizobium meliloti[22].

Fig.1 Relative molecular mass of standards (Lane 1) and purified ArsH (lane 2)
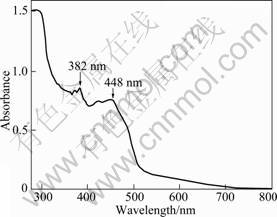
Fig.2 UV-vis absorbtion spectrum of purified ArsH
3.4 Activity assays of ArsH protein from E.coli BL21
The NADPH:FMN reductase activity of the purified ArsH from E. Coli BL21(DE3) pLysS was assayed. The enzymatic activity was determined through NADPH oxidation by monitoring the decrease in absorbance at 340 nm. From Fig.3, we knew that the optimal pH was 6.4 and the enzymatic activity remained high in an acid condition, but it lost a lot in alkali condition. When the reaction pool pH was decreased to 5.0, NADPH was unstable (data not shown). In conclusion, it was suggested that the ArsH protein cannot play a role in extreme acid environment and alkali environment.
3.5 Molecular structure modelling of ArsH protein
from At. ferrooxidans DX5
In the result of the FASTA search, the ArsH protein from Sinorhizobium meliloti (PDB code 2Q62) had a high level of sequence identity of 69% with the ArsH protein from At. ferrooxidans DX5, which should
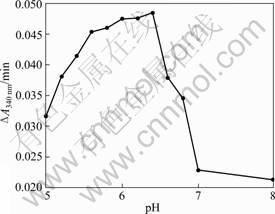
Fig.3 ArsH enzymatic activity
guarantee a relatively reliable homology structure building.
Based on the structure of 2Q62, through the procedures of homology modelling and molecular simulations, the final structure of the ArsH protein expressed from E. Coli BL21(DE3) pLysS was obtained.
The ProStat was used to calculate the percent of backbone Φ-Ψ angles within the allowed Ramachandran region. The result was that 89.5% of the Φ-Ψ angles in the model protein lay in the core region of the Ramachandran plot. For the X-ray structure of 2Q62, the percent of backbone Φ-Ψ angles was 90.5%. The analysis by ProStat also showed that there was no significant difference between the calculated values of the bond lengths and bond angles and those of the known proteins for the total residues. The results indicated that the modeled molecular structure was reliable.
4 Discussion
The arsH gene from At. ferrooxidans DX5 was amplified and successfully expressed in E.coli. The nucleotide sequence contained an open reading frame encoding a 237 amino acids protein, of which 100 were non-polar, 76 polar, 31 basic and 30 acidic amino acids. The percentages were calculated as 42.19% non-polar, 32.07% polar, 13.08% basic and 12.66% acidic residues. The deduced amino acid sequence was hydrophilic in nature, which was characterless of transmembrane protein. In addition, the transmembrane helixes of ArsH were analyzed with TMHMM Severs V2.0 (Fig.4) (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and the ArsH protein expressed from E.coli BL21 had no transmembrane helixes. The probability that the N-terminal was inside the cytoplasmic membrane was less than 0.025, strongly suggesting that the N-terminal was possible to be outside the cytoplasm, which may be functionally related to the arsenite- or arsenate- resistance.

Fig.4 TMHMM posterior probabilities prediction for transmembrane helixes of ArsH protein from E.coli BL21.
We all have known that the reduction of arsenate to arsenite is required for arsenide detoxification by bacterial cells[23]. The arsC gene encodes an arsenate reductase that reduces less toxic arsenate [As(V) (HAsO42-)] to the more toxic arsenite [As(III) (H3AsO3)], which occurs in the cytoplasmic side of the membrane, then the arsenite is pumped out of the cell through the action of arsB[12]. BUTCHER et al[4] provided genetic evidence that the reduction of arsenate by arsC in At. ferrooxidans is coupled to thioredoxin. In this study, NADPH appears to be the reductant and FMN acts as an electron transporter. On one hand, FMN can obtain hydrogen from NADPH and is reduced to FMNH2; on the other hand, the reduced FMNH2 is oxidized by O2 and returns to FMN state. This biochemical reaction occurs at the exterior of the membrane. Finally, electrons and protons react with O2 and form hydrogen peroxide[21-22] at outside of the membrane. The transformation rate of As(III) has been found to approach that of As(V) when pre-aerated water samples were treated with 7.5 mL/L H2O2 solution[24]. H2O2, the final product of the ArsH reaction, can accumulate and provide an oxidation potential to oxidize arsenite [As(III)] back to arsenate [As(V)] at the exterior of the membrane, which would convert toxic arsenite back to a less toxic form[25].
Compared with amino acid sequences in the NCBI database using the BLAST search tool, two homologous proteins with known functions were found that were over 76% identity to the ArsH protein from At. ferrooxidans DX5. The homologous proteins are the ArsH protein from Pseudomonas stutzeri A1501 (Accession No. NC_009434) and a NADPH-dependent FMN reductase from Pseudomonas mendocina ymp (Accession No. NC_009439). Using the NCBI website, a functional
domain was found in the ArsH protein of At. ferrooxidans DX5. This is a single domain, named FMN_red by NCBI, with a resolved crystalloid structure and is conjectured to represent an NADPH-dependent FMN reductase. Using the structure of the ArsH flavoprotein from Sinorhizobium meliloti[22], the putative structure of the ArsH flavoprotein from At. ferrooxidans DX5 was simulated (Fig.5). Like T1501 from P. aeruginosa[26] and the ArsH flavoprotein from Sinorhizobium meliloti[22], the ArsH flavoprotein from At. ferrooxidans DX5 also lacks a six-residue flavodoxin fingerprint motif (T/S)XTGXT. The putative FMN binding site on ArsH (GSNRECSYS) from At. ferrooxidans DX5 is similar to that of the T1501 FMN binding site (GSLRSGSYN) and of the ArsH protein from Sinorhizobium meliloti (GSLRTVSYS). Ser-39, Arg-41, Ser-44, Tyr-45 and Ser-46 comprise a sulfate interaction site that probably occupies a ribityl phosphate binding site, while residues 103-107 interact with the isoalloxazine ring of FMN.
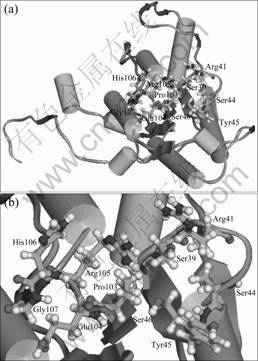
Fig.5 Overall structure (a) and local structure (b) of ArsH protein from At. ferrooxidans DX5
In conclusion, we reported the first expression in E. coli of the arsH gene from At. ferrooxidans DX5. The E.coli expressed ArsH protein was a flavoprotein containing FMN as the cofactor. The protein was shown to be structurally homologous to NADPH-dependent FMN reductases. The enzyme was demonstrated to have NADPH-dependent FMN reductase activity and the optimal pH for the enzyme activity was 6.4.
References
[1] BHATTACHARJEE H, ROSEN B P. Arsenic metabolism in prokaryotic and eukaryotic microbes [M]//NIES D H, SILVER S, Eds. Molecular Microbiology of Heavy Metals. Heidelberg, New York, Springer-Verlag, 2007: 388-390.
[2] MUKHOPADHYAY R, ROSEN B P. Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase [J]. FEMS Microbiol Lett, 1998, 168: 127-136.
[3] PATRA M, BHOWMIK N, BANDOPADHYAY B, SHARMA A. Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance [J]. Environ Exp Bot, 2004, 52: 199-223.
[4] BUTCHER B G, DEANE S M, RAWLINGS D E. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli [J]. Appl Environ Microbiol, 2000, 66: 1826-1833.
[5] BUTCHER B G, RAWLINGS D E. The divergent chromosomal ars operon of Acidithiobacillus ferrooxidans is regulated by an atypical ArsR protein [J]. Microbiology (Reading Engl), 2002, 148: 3983-3992.
[6] MUKHOPADHYAY R, ROSEN B P, PHUNG L T, SILVER S. Microbial arsenic: From geocycles to genes and enzymes [J]. FEMS Microbiol Rev, 2002, 26: 311-325.
[7] PRITHIVIRAJSINGH S, MISHRA S K, MAHADEVAN A. Detection and analysis of chromosomal arsenic resistance in Pseudomonas fluorescens strain MSP3 [J]. Biochem Biophys Res Commun, 2001, 280: 1393-1401.
[8] ROSEN B P. Families of arsenic transporters [J]. Trends Microbiol, 1999, 7: 207-212.
[9] SALTIKOV C W, CIFUENTES A, VENKATESWARAN K, NEWMAN DK. The ars detoxification system is advantageous but not required for As(V) respiration by the genetically tractable Shewanella species strain ANA-3 [J]. Appl Environ Microbiol, 2003, 69: 2800-2809.
[10] SILVER S, PHUNG L T. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic [J]. Appl Environ Microbiol, 2005, 71: 599-608.
[11] RAWLINGS D E. High level arsenic resistance in bacteria present in biooxidation tanks used to treat gold-bearing arsenopyrite concentrates: A review [J]. Transactions of Nonferrous Metals Society of China, 2008, 18: 1311-1318.
[12] TUFFIN I M, de GROOT P, DEANE S M, RAWLINGS D E. An unusual Tn21-like transposon containing an ars operon is present in highly arsenic resistant strains of the biomining bacterium Acidithiobacillus caldus [J]. Microbiology (Reading Engl), 2005, 151: 3027-3039.
[13] NEYT C, IRIARTE M, THI V H, CORNELIS G R. Virulence and arsenic resistance in Yersiniae [J]. J Bacteriol, 1997, 179: 612-619.
[14] NAVARRO C A, ORELLANA L H, MAURIACA C, JEREZ C A. Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper [J]. Appl Environ Microbiol, 2009, 75: 6102-6109.
[15] VORONTSOV I I, MINASOV G, BRUNZELLE J S, SHUVALOVA L, KIRYUKHINA O, COLLART F R, ANDERSON W F. Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity [J]. Protein Sci, 2007, 16: 2483-2490.
[16] WU Xue-ling, DING Jian-nan, GAO Jian, LIU Xin-xing, QIU Guan-zhou. Isolation and identification of metal-resistant iron-oxidizing bacteria [J]. Miner Metall Process, 2007, 24: 57-60.
[17] SODEOKA M, LARSON C, CHEN L, LAND W, VERDINE G. A multifunctional plasmid for protein expression by ECPCR: overproduction of the p50 subunit of NF-KB [J]. Bioorg Med Chem Lett, 1993, 3: 1095-1100.
[18] INOUYE S, SAHARA Y. Identification of two catalytic domains in a luciferase secreted by the copepod Gaussia princes[J]. Biochem Biophys Res Commun, 2008, 365: 96-101.
[19] INOUYE S, SASAKI S. Overexpression, purification and characterization of the catalytic component of oplophorus luciferase in the deep-sea shrimp, oplophorus gracilirostris[J]. Protein Expr Purif, 2007, 56: 261-268.
[20] LAEMMLI U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4[J]. Nature, 1970, 227: 680-685.
[21] BRUCHHAUS I, RICHTER S, TANNICH E. Recombinant expression and biochemical characterization of an NADPH: flavin oxidoreductase from Entamoeba histolytica[J]. Biochem J, 1998, 330: 1217-1221.
[22] YE J, YANG H, ROSEN B P, BHATTACHARJEE H. Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti[J]. FEBS Lett, 2007, 581: 3996-4000.
[23] APOSHIAN H V, ZAKHARYAN R A, AVRAM M D, SAMPAYOREYES A, WOLLENBERG M L. A review of the enzymology of arsenic metabolism and a new potential role of hydrogen peroxide in the detoxication of the trivalent arsenic species[J]. Toxicol Appl Pharmacol, 2004, 198: 327-335.
[24] YUAN Tao, LUO Qi-fang. Study on the removal of arsenite from dispersed drinking water[J]. Journal of Hygiene Research, 2001, 30: 70-71, 85. (in Chinese)
[25] MAZUELOS A, PALENCIA I, ROMERO R, RODRIGUEZ G, CARRANZA F. Ferric iron production in packed bed bioreactors: Influence of pH, temperature, particle size, bacterial support material and type of air distributor[J]. Miner Eng, 2001, 14: 507-514.
[26] AGARWAL R, BONANNO J B, BURLEY S K, SWAMINATHAN S. Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal[J]. Acta Crystallogr D: Biol Crystallogr, 2006, 62: 383-391.
(Edited by YANG Bing)
Foundation item: Project(50621063) supported by the National Natural Science Foundation of China; Project(2004CB619201) supported by the National Basic Research Program of China
Corresponding author: QIU Guan-zhou; Tel: +86-731-88879212; E-mail: qgzcsu@yahoo.cn
DOI: 10.1016/S1003-6326(09)60406-4