Production and characterization of alkaline extracellular lipase from newly isolated strain Aspergillus awamori HB-03
来源期刊:中南大学学报(英文版)2011年第5期
论文作者:夏金兰 黄斌 聂珍媛 王威
文章页码:1425 - 1433
Key words:alkaline lipase; Aspergillus awamori; purification; characterization
Abstract:
A strain HB-03 to produce alkaline extracellular lipase was isolated from oil-rich soil samples and identified as Aspergillus awamori. The growth conditions and nutritional factors for lipase production by strain HB-03 were optimized, and the maximum lipase production of (45.9±2.3) U/mL was obtained at 30 ℃ and pH 7.0 after 36 h using olive oil (1%) and sucrose (0.5%) as carbon sources and combination of peptone (2%), yeast extract (0.5%) and ammonium sulfate (0.1%) as nitrogen sources. The lipase was purified to homogeneity with 10.6-fold, 18.84% yield and a specific activity of 1 862.2 U/mg using ammonium sulfate precipitation followed by SephadexG-75 gel filtration chromatography. The purified lipase with molecular mass of 68 ku was estimated by SDS-PAGE. The optimum pH and temperature for the purified lipase were found to be 8.5 and 40 ℃, respectively. The lipase kept more than 80% of activity in pH 7.0-10.0 and temperatures up to 45 ℃. The metal ions of Mn2+, Ba2+ significantly enhanced the lipase activity, whereas Cu2+, Fe3+ and Mg2+ strongly reduced the lipase activity. The Km and Vmax values of the purified enzyme for p-nitrophenyl palmitate were 0.13 mmol/L and 60.6 mmol/(L·min), respectively. The results show that this novel lipase has potential industrial applications.
J. Cent. South Univ. Technol. (2011) 18: 1425-1433
DOI: 10.1007/s11771-011-0857-5![]()
XIA Jin-lan(夏金兰), HUANG Bin(黄斌), NIE Zhen-yuan(聂珍媛), WANG Wei(王威)
School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China
? Central South University Press and Springer-Verlag Berlin Heidelberg 2011
Abstract: A strain HB-03 to produce alkaline extracellular lipase was isolated from oil-rich soil samples and identified as Aspergillus awamori. The growth conditions and nutritional factors for lipase production by strain HB-03 were optimized, and the maximum lipase production of (45.9±2.3) U/mL was obtained at 30 °C and pH 7.0 after 36 h using olive oil (1%) and sucrose (0.5%) as carbon sources and combination of peptone (2%), yeast extract (0.5%) and ammonium sulfate (0.1%) as nitrogen sources. The lipase was purified to homogeneity with 10.6-fold, 18.84% yield and a specific activity of 1 862.2 U/mg using ammonium sulfate precipitation followed by SephadexG-75 gel filtration chromatography. The purified lipase with molecular mass of 68 ku was estimated by SDS-PAGE. The optimum pH and temperature for the purified lipase were found to be 8.5 and 40 °C, respectively. The lipase kept more than 80% of activity in pH 7.0-10.0 and temperatures up to 45 °C. The metal ions of Mn2+, Ba2+ significantly enhanced the lipase activity, whereas Cu2+, Fe3+ and Mg2+ strongly reduced the lipase activity. The Km and Vmax values of the purified enzyme for p-nitrophenyl palmitate were 0.13 mmol/L and 60.6 mmol/(L?min), respectively. The results show that this novel lipase has potential industrial applications.
Key words: alkaline lipase; Aspergillus awamori; purification; characterization
1 Introduction
Lipase (triacylglycerol acylhydrolase, EC 3.1.1.3), which is one kind of the most important industrial enzymes, can catalyze the hydrolysis of triglycerides into fatty acids and glycerol at the interface between the aqueous and organic phases [1-2]. Lipases were widely found in many kinds of animals, plants and microorganisms [3]. However, microbial lipases have received much more attention than the others, because of their availability, high yield, good activities, stability and enormous potential applications in various industries [4-5].
It is well known that the Aspergillus species can produce a variety of enzymes including lipase, hemicellulase, pectinase, phytase, glycosidase, amylase, cellulase, protease and peptidase [6]. Because of their inherent metabolic diversity, they can grow on a wide range of substrates and show much more advantages in industrial and catalytic applications. In recent years, a number of reports on lipases from Aspergillus species have been published [2, 4, 7]. Generally, lipases from Aspergillus strains are active in acidic conditions (pH 4.0-7.0) and in the temperature range of 40-50 °C [7]. Nevertheless, alkaline lipases are more commercially important than acidic lipases [8], but little research has been done on extracellular alkaline lipase from Aspergillus species.
In this work, an extracellular alkaline lipase- producing strain was isolated and identified. Furthermore, the effects of culture conditions and nutritional factors on lipase production of that strain, and the biochemical characterization of the lipase were investigated.
2 Experimental
2.1 Samples and culture media
In order to gain alkaline lipase-producing microorganisms, soil samples were collected from meat processing combine and various oil spilling sites in Changsha, China.
Enrichment medium contained (per liter): peptone 20 g, yeast extract 5 g, MgSO4·7H2O 0.5 g, olive oil 30 mL and pH is 8.0. Tributyrin agar plates contained (per liter): (NH4)2SO4 1 g, K2HPO4 1 g, KCl 0.5 g, MgSO4·7H2O 0.5 g, agar 15 g, tributyrin 10 mL, bromocresol purple 0.01 g, and pH is 8.0. Fermentation medium contained (per liter): peptone 40 g, sucrose 5 g, (NH4)2SO4 1 g, MgSO4·7H2O 1 g, KH2PO4 1 g, olive oil 10 mL, and pH is 6.0.
The strains were cultured in 250 mL Erlenmeyer flasks containing 50 mL of liquid culture medium. All the culture media were sterilized at 115 °C for 30 min before use.
2.2 Isolation and screening of lipase-producing fungi
All the samples were cultivated on enrichment medium and incubated at 28 °C for 2 d, with 3 parallel experiments. Then, the strains were subjected to purification by serial dilution, and cultivated on tributyrin agar plates at 30 °C for 5 d, and lipase production was identified as a clear zone around colonies.
2.3 Identification of strain HB-03 using 18S rDNA gene sequence
The identification of lipase-producing fungi was fulfilled by 18S rDNA sequence analysis[9]. The 18S rDNA sequence of strain HB-03 was amplified by PCR with forward primer 5′-CCTGGTTGATCCTGCCAG-3′ and reverse primer 5′-TTGATCCTTCTGCAGGTTCA- 3′. The PCR reactions were carried out as follows: one initial cycle at 95 °C (5 min), followed by 34 cycles of 95 °C (1 min); annealing at 55 °C (1 min); 72 °C (1.5 min); and ended with incubation at 72 °C for 10 min. The amplified product was electrophoresed on a 1.0% agarose gel. The fragment of the interesting band was excised from the gel and purified with Rapid Recovery Kit (Gel), followed by sequencing (Sangon Shanghai, China). Homological sequence alignment of the 18S rDNA sequence with other origin from Genbank was performed by using Clustal X 1.8 software, and then phylogenetic tree was constructed with MEGA 3.1 software.
2.4 Lipase production
0.5 mL spores suspension (1.0×106 mL-1) was prepared by washing 7 days-old potato dextrose agar (PDA) slants with sterile distilled water, which was added into 250 mL Erlenmeyer flasks with 50 mL fermentation medium, and incubated at 28 °C, 180 r/min for 48 h in a rotary shaker. At the end of incubation period, cells were harvested by centrifugation (10 000 g, 10 min at 4 °C), and the cell-free supernatant was used for determining the crude lipase activity.
2.5 Enzyme assay
The lipase activity was determined using p-nitrophenyl palmitate (p-NPP) as substrate according to WINKLER and STUCKMANN [10] with some modification. In brief, solution A consisted of 90 mg p-NPP dissolved in 30 mL HPLC grade iso-propanol, then stored at -20 °C immediately, and solution B contained 0.4% (v/v) Triton X-100 and 0.1% (w/v) gum arabic dissolved in 0.05 mol pH 8.0 Tris-HCl buffer. The reaction mixture with 0.5 mL solution A and 4.5 mL solution B was pre-incubated for 5 min at 37 °C. Then, 0.05 mL enzyme was added into the reaction mixture and incubated for 10 min at 37 °C in a shaking water bath, which was terminated by the addition of ethanol. And the inactivated enzyme, which was heated at 100 °C for 5 min, was used as control following the same procedure. Finally, the lipase activity was detected at 410 nm with spectrophotometer. One unit of lipase was defined as the amount of enzyme, which liberates 1.0 μmol p-nitrophenol (p-NP) from p-NPP per minute under standard assay conditions.
2.6 Effect of initial pH, temperature and time of incubation on lipase production
In order to find out the optimum initial pH and temperature of incubation for the lipase production, the experiments were carried out with the initial pH values in the range of 6.0-10.0 and temperatures were detected at 24, 26, 28, 30, 32 and 34 °C. To find out the relationship between the lipase production and the cultivation time, the lipase production curve against time was drawn every 6 h for 72 h.
2.7 Purification of enzyme
The lipase was purified by two steps, and all operations were carried out at 4 °C. After fermentation for 36 h in optimized fermentation medium, culture broth was filtered and centrifuged at 10 000g for 10 min at 4 °C to obtain the cell-free supernatant. And the supernatant was concentrated with ammonium sulfate precipitation at 90% of saturation with continuous stirring and left overnight. The protein precipitate was collected by centrifugation (10 000g, 20 min at 4 °C) and dissolved in minimal amount of 20 mmol/L PBS buffer (pH 7.2). Then, concentrated sample was loaded on a Sephadex G-75 column (1.6 cm × 50 cm) pre-equilibrated with 20 mmol/L PBS buffer (pH 7.2). The column was washed with the same buffer at a flow rate of 0.5 mL/min. Finally, the active fractions were pooled and concentrated by polyethylene glycol 8000 (PEG 8000), and the purified lipase was stored at -20 °C for further studies.
2.8 Determination of protein and molecular mass
The concentration of protein was determined by the method of BRADFORD et al[11]. And the relative mass of lipase was assayed by SDS-PAGE as described by LAEMMLI [12].
2.9 Characterization of purified lipase
The effect of pH on enzyme activity was determined by measuring the enzyme activity at pH from 7.0 to 11.0. The buffers used were 0.05 mol/L Tris-HCl (pH 7.0-9.0), 0.05 mol/L Gly-NaOH (pH 9.0-11.0). The pH stability was obtained by measuring the relative activity of enzyme after 60 min of pre-incubation in 0.05 mol/L buffers of various pH values (7.0-11) at 37 °C.
The effect of temperature on the activity of the lipase was determined by assaying the relative enzyme activity at different temperatures from 30 to 70 °C with Tris-HCl buffer (0.05 mol/L, pH 8.5). The thermal stability of lipase was measured at different temperatures from 30 to 70 °C for 60 min with pH 8.5, 0.05 mol/L Tris-HCl buffer.
The effects of various metal ions (1 mmol/L and 10 mmol/L ) such as Ca2+, Mg2+, Zn2+, Mn2+, Cu2+ and Fe3+ were determined by being pre-incubated with the lipase for 60 min and then the residual lipase activity was measured.
2.10 Determination of kinetic parameters
To obtain Km and Vmax of the purified lipase, kinetic experiments were performed under the standard lipase assay using various concentrations of p-NPP as substrate. The values of kinetic parameters, Km and Vmax, were determined from Lineweaver–Burk plots.
3 Results and discussion
3.1 Isolation and screening of strains with lipase activity
Eighteen of total 58 strains isolated could form clear zone around the colonies on the tributyrin agar plates (results not shown). The results indicated that lipase activity of strain HB-03 was the highest, which grew better on the triacylglycerol plates and the diameter of zone hydrolysis was 18 mm after 5 d cultivation. Therefore, strain HB-03 was chosen for lipase-producing strain and used for the further studies.
3.2 Morphological characteristics and identification of strain HB-03
Strain HB-03 can grow in a broad temperature range of 20-45 °C and a wide pH range of 4.0-10.0, and for a longer incubation time, more spores would form according to optical microscope and colony color observation. When being cultured at 30 °C and pH 7.0 on a PDA plate for 7 d, the colony of strain HB-03 was large, white and villiform with radial circumference, black color on the surface and pale yellow on the back, which demonstrated the formation of spores (as shown in Fig.1).
The 18S rDNA of strain HB-03 was amplified, sequenced, and submitted to GenBank (Accession No.GU226429). The sequencing result showed that the amplified 18S rRNA was 1733 bp. Then, a comparative analysis was performed by Clustal X 1.8 and MEGA 3.1 software, which indicated that 18S rDNA gene sequence from strain HB-03 had a significant identity to Aspergillus awamori strain 07-02 (as shown in Fig.2). In terms of 18S rDNA gene sequence, morphological characteristics and phylogenetic tree construction, strain HB-03 was identified as Aspergillus awamori.
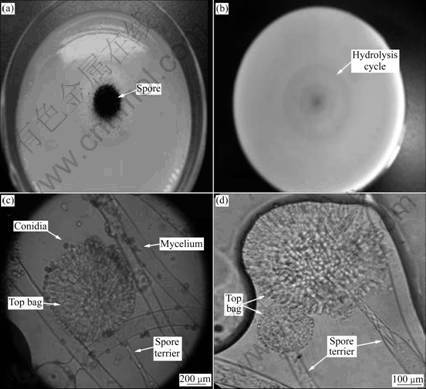
Fig.1 Morphological features of strain HB-01: (a) Colony formed on PDA for 5 d; (b) Colony formed on solidified selected medium for 5 d; (c), (d) Microscopic morphologies
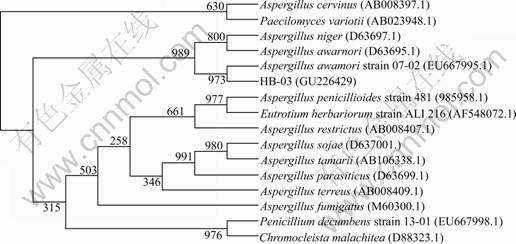
Fig.2 Phylogenetic tree for strain HB-03 and relative strains based on 18S rDNA gene sequence
3.3 Effect of initial pH of culture medium on lipase production
It has been noted that the initial pH of culture medium plays an important role in the cell growth and enzyme production [13]. The results of pH studies revealed that strain HB-03 showed the highest lipase activity (43.7 U/mL) at initial pH 7.0 of the production medium (as shown in Fig.3). It was reported the genus of Aspergillus can grow in a wide range of pH for lipase production. The best pH for lipase production from Aspergillus terreus was 9.0 [14]. On the contrary, lipase production by Aspergillus sp. was the maximum at pH 5.5 [15].
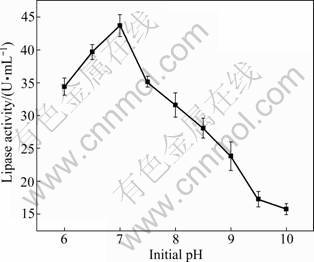
Fig.3 Effect of initial pH of culture medium on lipase production
3.4 Effect of incubation temperature on lipase production
It is well known that temperature plays an important role in the biosynthesis of lipase by microorganisms. Generally, the optimum temperature for lipase production from genus of Aspergillus ranges from 28 to 40 °C. Similar to most strains of Aspergillus species, the results on the effect of temperature showed that the lipase activity reached the highest when the strain was grown at 30 °C (Fig.4). It was the same as the report of Aspergillus sp. which was also incubated at 30 °C, and the maximum lipase production was gained [15]. However, the best temperature for lipase production from Aspergillus terreus was 37 °C [14].
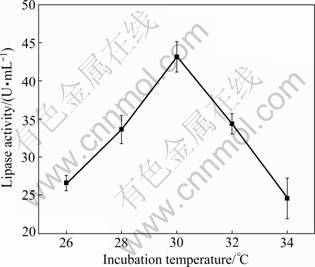
Fig.4 Effect of incubation temperature on lipase production
3.5 Effect of inducers on lipase production
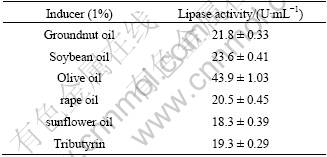
The effect of different inducers on lipase production was studied. As the results (Table 1) indicated, the maximum lipase activity ((43.9±1.03) U/mL) was obtained with olive oil as the inducer. Groundnut oil and soybean oil were also proved to be fine inducers for lipase production of A. awamori HB-03. Similarly, til and olive oils were used for lipase production by A. niger NCIM1207 and the maximum yields of lipase activity were obtained [16]. However, inducers except olive oil cannot enhance lipase production of A. niger [17].
Table 1 Effect of inducers on lipase production
3.6 Effect of carbon sources on lipase production
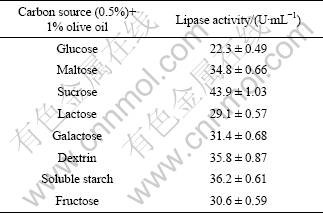
As we know, different carbon sources have different effects on the enzyme production. Glucose, maltose, sucrose, lactose, galactose, dextrin, soluble starch and fructose were tested as carbon sources at 0.5% in media supplemented with 1% olive oil. As Table 2 indicated, all carbon sources could enhance the lipase production except glucose. The highest lipase production ((43.9±1.03) U/mL) was observed with sucrose as the carbon source. However, sucrose was reported to have no significant influence on the lipase production by A. niger [17]. Generally, microorganisms can produce high yields of lipase when carbon sources are used. Bacillus cereus C7 produced lipase when the medium contained 2% starch as carbon source [18]. Fructose and glucose were reported to be the best carbon sources for lipase production by Amycolatopsis mediterranei DSM 43304 and Candida cylindracea, respectively [19-20].
Table 2 Effect of carbon sources on lipase production
3.7 Effect of nitrogen sources on lipase production
In general, nitrogen source is one of the most critical factors for lipase production. It can be observed from the results that the lipase activity ((44.8±1.14) U/mL) of strain HB-01 was obtained with 2% peptone and 0.5% yeast extract as organic nitrogen sources in the medium (Table 3). Similarly, many reports showed that peptone and yeast extract significantly enhanced lipase production [20-21]. It can also be seen from the results that (NH4)2SO4 was the best inorganic nitrogen source which could significantly increase the lipase production of strain HB-01. But urea was found to inhibit the lipase production. However, urea was reported to enhance lipase production from Rhodotolura glutinis [22].
Table 3 Effect of nitrogen sources on lipase production

3.8 Time courses of lipase production and cell growth
The time courses of lipase production and cell growth were investigated at the optimal culture parameters. As Fig.5 showed, the growth of strain HB-03 got into the exponential phase after 18 h and reached stationary phase after 42 h. It could also be observed from the results that alkaline lipase production increased quickly at the exponential phase and achieved the maximum yield at the end of the exponential phase, then declined rapidly. The optimum lipase production of (45.9±2.3) U/mL was observed during the late exponential phase and beginning of the stationary phase at 36 h. It was reported that the highest lipase production of Rhizopus rhizopodiformis S1 was obtained after 72 h cultivation, then the lipase production decreased, companied with the biomass increasing [23].
3.9 Purification of lipase
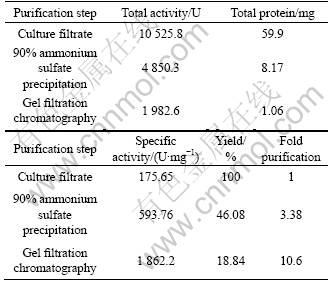
The result of the purification of extracellular lipase from A. awamori HB-03 is shown in Table 4. After two steps of purification, the lipase exhibited 1 862.2 U/mg specific activity and 10.6-fold purification with a yield of 18.84%.
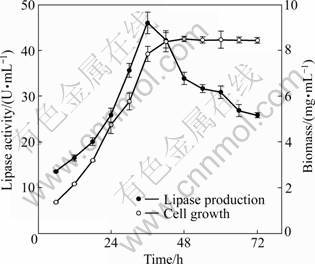
Fig.5 Time course of lipase production and cell growth by using modified medium at optimal culture conditions
Table 4 Summary of purification of extracellular lipase from A. awamori HB-03
The purified lipase showed one protein band on SDS-PAGE, and the molecular mass was 68 ku estimated by SDS-PAGE (Fig.6). The previous studies showed that the molecular masses of most Aspergillus lipases were in the range of 29-70 ku [7, 24]. The lipase from Aspergillus oryzae was a monomeric protein, which had a molecular mass of 41 ku determined by SDS-PAGE and 39 ku by gel filtration [25]. And the lipase from Aspergillus niger had a molecular mass of 35.5 ku by SDS-PAGE [26].
3.10 Effect of pH on lipase activity and stability
The effect of pH on the lipase activity is shown in Fig.7. The lipase was active over a broad pH range, and the optimum for the lipase activity was observed at pH 8.5. The optimum alkaline pH is different from other Aspergillus lipases, for example, the lipase produced by A. niger and A. niger NCIM 1207, both exhibited optimum pH of 2.5 [7, 16]. However, similar results were observed in lipase from A. carneus which presented optimum alkaline pH of 9.0 [27].

Fig.6 Molecular mass determination of lipase by SDS-PAGE (Lane M: Maker proteins; Lane 1: Purified lipase)
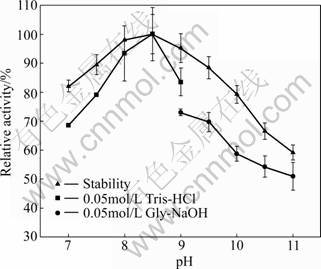
Fig.7 Effect of pH on lipase activity and stability
In order to determine the pH stability of the enzyme, the lipase was pre-incubated in buffers between pH 7.0 and 11.0 for 60 min. It was found that the highest stability of the lipase was at pH 8.0-9.0 where the activity decreased only by 5%, while the pH stability of the lipase decreased rapidly at pH 10.0-11.0. It was reported that lipases from A. niger and A. niger NRRL3 were unstable at alkaline pH [28-29].
3.11 Effect of temperature on lipase activity and stability
The effect of temperature on lipase activity of strain HB-03 was determined after being incubated at temperatures between 30 and 70 °C in Tris-HCl buffer (0.05 mol/L, pH 8.5) for 10 min. As Fig.8 showed, the optimum temperature for the lipase activity was about 40 °C, which was similar to that reported for the lipase of A. carneus [27]. In contrast with these results, the optimum temperature of the partially purified lipase from A. niger NRRL3 and A. niger was 60 °C and 25 °C, respectively [28-29]. It can also be seen from the results that the lipase retained almost 100% of activity at 30-45 °C after 60 min incubation and then the lipase activity decreased rapidly.

Fig.8 Effect of temperature on lipase activity and stability
3.12 Effect of metal ions on lipase activity
The effects of different metal ions on the lipase activity were tested by assaying the activity after incubation for 10 min at 40 °C and pH 8.5 with 1 mmol/L or 10 mmol/L of the metal ions (Table 5). 1 mmol/L Cu2+ and Fe3+ strongly inhibited the lipase activity to 51% and 56%, respectively. In contrast, 1 mmol/L Mn2+, Ba2+ significantly promoted the lipase activity to 122% and 135%, respectively. Interestingly, Zn2+ increased the lipase activity at 1 mmol/L, but inhibited the lipase activity at 10 mmol/L. When the concentrations were 10 mmol/L, the enzyme activity was slightly inhibited by Zn2+ and Ca2+. At the same time, the lipase activities were enhanced by 1.6 times and 0.8 times with addition of 10 mmol/L of Mn2+ and Ba2+, respectively. Similarly, it was reported by YAMAMOTO and FUJIWARA [30], the lipase activity was strongly inhibited by Ca2+. However, lots of reports showed that the lipase activities were enhanced in the presence of Ca2+ [18, 31].
3.13 Effect of effectors on lipase activity
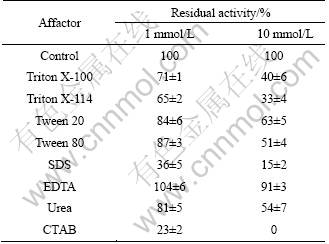
The effects of different factors on lipase were assessed (Table 6). It can be observed that EDTA slightly enhanced the enzyme activity at the concentration of 1 mmol/L, whereas the lipase was inhibited in the presence of SDS, Triton X-100, Triton X-114, Tween 20, Tween 80, urea. And the lipase was strongly inhibited by CTAB losing 80% and 100% of its activity at the concentrations of 1 mmol/L and 10 mmol/L, respectively. It was reported that 0.2% SDS had weak stimulatory effect on lipase activity [18], but no effect was observed on lipase stability of A. mediterranei DSM 4334 at the concentration of 1 mmol/L [19]. Same as the reports on lipases from S. fradiae var. k11, A. mediterranei DSM 4334 and S. rimosus, EDTA had little effect on the lipase activity of strain HB-03 [19, 32-33].
Table 5 Effect of metal ions on lipase activity
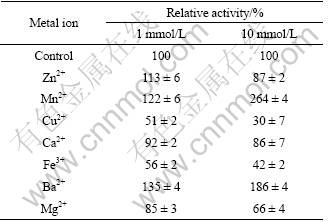
Table 6 Effect of affectors on lipase activity
3.14 Kinetic parameters of lipase from strain HB-03
As Lineweaver-Burk plot showed, the Km and Vmax values of the lipase for p-NPP (Fig.9) were 0.13 mmol/L and 60.6 mmol/(L?min), respectively. For a lipase from Pseudomonas cepacia, the Km and Vmax values were 12 mmol/L and 30 μmol/(L?min), respectively, when the substrate was p-NPP[34]. And the Km and Vmax values of Pseudomonas aeruginosa SRT 9 with the p-NPP as substrate were 0.037 mmol/L and 188.6 mmol/(L?min), respectively [35]. These results indicated that the purified lipase from Aspergillus awamori HB-03 displayed very high affinity for p-NPP.

Fig.9 Lineweaver-Burk plot for Km and Vmax values of lipase in presence of various concentrations of p-NPP
4 Conclusions
1) A lipase-producing strain HB-03 from oil-rich soil samples from Changsha, China, was isolated and identified as Aspergillus awamori.
2) The growth conditions and nutritional factors for lipase production by strain HB-03 were optimized, and the maximum lipase production of (45.9±2.3) U/mL was obtained at 30 °C and pH 7.0 after 36 h incubation using olive oil (1%) and sucrose (0.5%) as carbon sources and combination of peptone (2%), yeast extract (0.5%) and ammonium sulfate (0.1%) as nitrogen sources.
3) The extracelluar lipase secreted by the strain Aspergillus awamori HB-03 was precipitated by 90% ammonium sulfate and purified by Sephadex G-75 gel filtration chromatography. Purifications of 10.6 fold and 18.84% yield over the initial material were achieved. The specific activity of the lipase was 1 862.2 U/mg. The molecular mass of the purified lipase was 68 ku.
4) The optimum pH and temperature for the purified lipase were found to be 8.5 and 40 °C, respectively. The lipase kept almost 90% of activity in pH 7.0-10.0 and temperatures up to 45 °C. The metal ions of Mn2+, Ba2+ significantly enhanced the lipase activity, whereas Cu2+, Fe3+ and Mg2+ strongly reduced its activity. The Km and Vmax value of the purified enzyme for p-nitrophenyl palmitate were 0.13 mmol/L and 60.6 mmol/(L?min), respectively.
References
[1] MACRAE A, HAMMOND R. Present and future applications of lipases [J]. Biotechnology and Genetic Engineering Reviews, 1985, 3: 193-217.
[2] KAUSHIK R, SARAN S, ISAR J, SAXENA R. Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus [J]. Journal of Molecular Catalysis B: Enzymatic, 2006, 40: 121-126.
[3] SZTAJER H, MALISZEWSKA J. Production of exogenous lipases by bacteria, fungi, and actinomycetes [J]. Enzyme and Microbial Technology, 1988, 10: 492-497.
[4] ELLAIAH P, PRABHAKAR T, RAMAKRISHNA B, TALEB A, ADINARAYANA K. Production of lipase by immobilized cells of Aspergillus niger [J]. Process Biochemistry, 2004, 39: 525-528.
[5] BJRKLING F, GODTFREDSEN S, KIRK O. The future impact of industrial lipases [J]. Trends in Biotechnology, 1991, 9: 360-363.
[6] MACCABE A, OREJAS M, TAMAYO E, VILLANUEVA A, RAM?N D. Improving extracellular production of food-use enzymes from Aspergillus nidulans [J]. Journal of Biotechnology, 2002, 96: 43-54.
[7] MHETRAS N, BASTAWDE K, GOKHALE D. Purification and characterization of acidic lipase from Aspergillus niger NCIM 1207 [J]. Bioresource Technology, 2009, 100: 1486-1490.
[8] CHEN Shu-jen, CHENG Chu-yuan, CHEN Teh-liang. Production of an alkaline lipase by Acinetobacter radioresistens [J]. Journal of Fermentation and Bioengineering, 1998, 86: 308-312.
[9] MOON S, KIM W. Phylogenetic position of the Tardigrada based on the 18S ribosomal RNA gene sequences [J]. Zoological Journal of the Linnean Society, 2008, 116: 61-69.
[10] WINKLER U, STUCKMANN M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens [J]. Journal of Bacteriology, 1979, 138: 663-670.
[11] BRADFORD M. A rapid and sensitive method for the quantification of microgram quantities of protein using the principle of protein-dye release [J]. Analytical Biochemistry, 1976, 72: 248-254.
[12] LAEMMLI U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4 [J]. Nature, 1970, 227: 680-685.
[13] KUMAR C, TAKAGI H. Microbial alkaline proteases from a bioindustrial viewpoint [J]. Biotechnology Advances, 1999, 17: 561-594.
[14] GULATI R, SAXENA R, GUPTA R, YADAV R, DAVIDSON W. Parametric optimisation of Aspergillus terreus lipase production and its potential in ester synthesis [J]. Process Biochemistry, 1999, 35: 459-464.
[15] CIHANGIR N, SARIKAYA E. Investigation of lipase production by a new isolate of Aspergillus sp. [J]. World Journal of Microbiology and Biotechnology, 2004, 20: 193-197.
[16] MAHADIK N, PUNTAMBEKAR U, BASTAWDE K, KHIRE J, GOKHALE D. Production of acidic lipase by Aspergillus niger in solid state fermentation [J]. Process Biochemistry, 2002, 38: 715-721.
[17] POKORNY D, FRIEDRICH J, CIMERMAN A. Effect of nutritional factors on lipase biosynthesis by Aspergillus niger [J]. Biotechnology Letters, 1994, 16: 363-366.
[18] DUTTA S, RAY L. Production and characterization of an alkaline thermostable crude lipase from an isolated strain of Bacillus cereus C7 [J]. Applied Biochemistry and Biotechnology, 2009, 159: 142-154.
[19] DHEEMAN D, FRIAS J, HENEHAN G. Influence of cultivation conditions on the production of a thermostable extracellular lipase from Amycolatopsis mediterranei DSM 43304 [J]. Journal of Industrial Microbiology and Biotechnology, 2010, 37: 1-17.
[20] MURALIDHAR R, CHIRUMAMILA R, MARCHANT R, NIGAM P. A response surface approach for the comparison of lipase production by Candida cylindracea using two different carbon sources [J]. Biochemical Engineering Journal, 2001, 9: 17-23.
[21] IIZUMI T, NAKAMURA K, FUKASE T. Purification and characterization of a thermostable lipase from newly isolated Pseudomonas sp. KWI-56 [J]. Agricultural and Biological Chemistry, 1990, 54: 1253-1258.
[22] PAPAPARASKEVAS D, CHRISTAKOPOULOS P, KEKOS D, MACRIS B. Optimizing production of extracellular lipase from Rhodotorula glutinis [J]. Biotechnology Letters, 1992, 14: 397-402.
[23] SAMAD M, SALLEH A, RAZAK C, AMPON K, YUNUS W, BASRI M. A lipase from a newly isolated thermophilic Rhizopus rhizopodiformis [J]. World Journal of Microbiology and Biotechnology, 1990, 6: 390-394.
[24] SHARMA R, CHISTI Y, BANERJEE U. Production, purification, characterization, and applications of lipases [J]. Biotechnology Advances, 2001, 19: 627-662.
[25] TOIDA J, KONDOH K, FUKUZAWA M, FUKUZAWA M, SEKIGUCHI J. Purification and characterization of a lipase from Aspergillus oryzae [J]. Bioscience, Biotechnology, and Biochemistry, 1998, 62: 759-763.
[26] HARIDASAN NAMBOODIRI V, CHATTOPADHYAYA R. Purification and biochemical characterization of a novel thermostable lipase from Aspergillus niger [J]. Lipids, 2000, 35: 495-502.
[27] SAXENA R, DAVIDSON W, SHEORAN A, GIRI B. Purification and characterization of an alkaline thermostable lipase from Aspergillus carneus [J]. Process Biochemistry, 2003, 39: 239-247.
[28] HATZINIKOLAOU D, MACRIS J, CHRISTAKOPOULOS P, KEKOS D, KOLISIS F, FOUNTOUKIDIS G. Production and partial characterisation of extracellular lipase from Aspergillus niger [J]. Biotechnology Letters, 1996, 18: 547-552.
[29] ADHAM N, AHMED E. Extracellular lipase of Aspergillus niger NRRL3: Production, partial purification and properties [J]. Indian Journal of Microbiology, 2009, 49: 77-83.
[30] YAMAMOTO K, FUJIWARA N. Purification and some properties of a castor-oil-hydrolyzing lipase from Pseudomonas sp. [J]. Agricultural and Biological Chemistry, 1988, 52: 3015-3021.
[31] HIOL A, JONZO M, DRUET D, COMEAU L. Production, purification and characterization of an extracellular lipase from Mucor hiemalis f. hiemalis [J]. Enzyme and Microbial Technology, 1999, 25: 80-87.
[32] ZHANG Yu-hong, MENG Kun, WANG Ya-ru, LUO Hui-ying, YANG Pei-long, SHI Peng-jun, WU Ning-feng, FAN Yun-liu, LI Jiang, YAO Bin. A novel proteolysis-resistant lipase from keratinolytic Streptomyces fradiae var. k11 [J]. Enzyme and Microbial Technology, 2008, 42: 346-352.
[33] ABRAMI M, LEI I, KORICA T, VITALE LJ, SAENGER W, PIGAC J. Purification and properties of extracellular lipase from Streptomyces rimosus* 1 [J]. Enzyme and Microbial Technology, 1999, 25: 522-529.
[34] PENCREAC’H G, BARATTI J. Hydrolysis of p-nitrophenyl palmitate in n-heptane by the Pseudomonas cepacia lipase: A simple test for the determination of lipase activity in organic media [J]. Enzyme and Microbial Technology, 1996, 18: 417-422.
[35] BORKAR P, BODADE R, RAO S, KHOBRAGADE C. Purification and characterization of extracellular lipase from a new strain: Pseudomonas aeruginosa SRT 9 [J]. Brazilian Journal of Microbiology, 2009, 40: 358-366.
(Edited by YANG Bing)
Received date: 2010-06-29; Accepted date: 2011-03-20
Corresponding author: XIA Jin-lan, Professor, PhD; Tel: +86-731-88836944; E-mail: jlxia@mail.csu.edu.cn

