
J. Cent. South Univ. (2018) 25: 1052-1062
DOI: https://doi.org/10.1007/s11771-018-3805-9
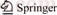
Panax ginseng-specific sequence characterized amplified region (SCAR) marker for testing medicinal products
JIANG Qiu-tao(蒋秋桃)1, 2, LIU Li(刘丽)2, XIAO Bing-yi(肖炳燚)2, LI Wen-li(李文莉)2,
LUO Hui-ming(罗晖明)2, NIE Ping(聂平)2, DING Ye(丁野)2, LI Jie(李洁)1, LI Wen-zhang(李文章)1
1. School of Chemistry and Chemical Engineering, Central South University, Changsha 410083, China;
2. Hunan Institute for Drug Control, Changsha 410001, China
 Central South University Press and Springer-Verlag GmbH Germany, part of Springer Nature 2018
Central South University Press and Springer-Verlag GmbH Germany, part of Springer Nature 2018
Abstract: To screen genetic polymorphisms of Panax ginseng, as well as those of Panax quinquefolium and Panax notoginseng, analysis of random amplified polymorphic DNA (RAPD) was performed using 120 random primers. Of the successful amplicons obtained, the Panax ginseng-specific RAPD marker C-12 was cloned into a TA vector and sequenced (GenBank access number KU553472). Based on the sequence analysis results, a pair of primers specific to C-12 was designed. Finally, a SCAR marker-based identification system for Panax ginseng was developed after optimization of the reaction conditions. Using this method, two positive bands were stably observed at 300 bp and 130 bp in 33 batches of Panax ginseng samples tested, while negative results were obtained for another 101 batches of samples, including Panax quinquefolium, Panax notoginseng, adulterants, and other medicinal herbs. Thus, we successfully developed a PCR-based method for rapid and effective identification of Panax ginseng, which can be effectively used for the protection and utilization of germplasm resources and identification of the origins of Panax ginseng samples.
Key words: Panax ginseng; random amplified polymorphic DNA (RAPD); sequence characterized amplified regions (SCAR); molecular identification
Cite this article as: JIANG Qiu-tao, LIU Li, XIAO Bing-yi, LI Wen-li, LUO Hui-ming, NIE Ping, DING Ye, LI Jie, LI Wen-zhang. Panax ginseng-specific sequence characterized amplified region (SCAR) marker for testing medicinal products [J]. Journal of Central South University, 2018, 25(5): 1052–1062. DOI: https://doi.org/10.1007/s11771-018- 3805-9.
1 Introduction
Ginseng drugs, including Ginseng (Panax ginseng C. A. Mey.), American ginseng (P. quinquefolium L.) and Notoginseng (P. notoginseng (Burk) F. H. Chen ex C. Chow & W. G. Huang) etc. are a group of valuable herbal medicines, which are widely used in Northern American and Asian countries, and have adaptogenic, restorative, immunomodulatory, vasodilatory, anti-inflammatory, antioxidant, anti-aging, anticancer, anti-fatigue, anti-stress, and anti-depressive effects in rodents and humans [1–6]. Driven by economic development and the on-going internationalization of traditional Chinese medicinal herbs, ginseng drugs have been utilized not only in traditional treatment but also in health-food products and foods. Since ginseng is obtained from the same part of different plant species belonging to the same genus and family, the above-mentioned three herbal medicines show highly similar internal structures and chemical compositions, making it very difficult to authenticate ginseng, particularly in single-drug or compound medicines. Unlawful practices such as substituting ginseng with other medicinal roots (e.g., the roots of Phytolacca acinosa Roxb. or Talinum paniculatum (Jacq.) Gaertn.) have become common in the market [7]. Counterfeit products do not have the claimed therapeutic effects and might even have toxic or adverse effects. The rapid adoption of DNA molecular marker-based technologies [8] to identify traditional Chinese herbal medicines and their decoction products has played an important role in authenticating medicinal products [9, 10].
Sequence characterized amplified region (SCAR) analysis [11], in which a single, genetically defined locus is identified by amplification of genomic DNA with a pair of specific oligonucleotide primers, is a highly sensitive and reliable molecular marker in various fields such as species identification, marker-assisted screening, and map-based gene cloning. In particular, SCAR is suitable for detecting differences in DNA among different samples. SCAR markers have the potential to replace RAPD (random amplified polymorphic DNA) and other DNA-based markers that are expensive, time-consuming, and tedious. SCAR tagging involves the recovery, cloning, and sequencing of specific RAPD fragments. This is followed by primer design based on the sequencing results, and the resulting primers are used to amplify genomic DNA by PCR. This method offers several benefits, including simplicity of operation, stable performance, and low cost [9, 12]. SCAR markers have been developed for authentication of various medicinal plants that can be easily adulterated. Currently, SCAR is used as a national standard for identification of pilose antler in Korean herbal pharmacopeia. ZHOU et al [13] obtained a SCAR marker specific for Polygonum capitatum (P. capitatum) using RAPD technology and successfully identified P. capitatum using specific SCAR fragments that were amplified with the constructed primers. A combination of RAPD and eastern blotting analyses using anti-ginsenoside Rb1 and Rg1 monoclonal antibodies by TANAKA et al [14] was used for the identification of P. notoginseng, P. quinquefolius and P. japonicas. JUNG et al [15] developed convenient and reliable chloroplast genome-derived DNA markers for authentication of Korean and American ginseng in commercial processed products. The developed markers were successfully applied to evaluating the original species from various processed ginseng products purchased from markets in Korea and China. High-throughput application of this marker system will eradicate illegal trade and promote confident marketing for both species to increase the value of Korean as well as American ginseng in Korea and worldwide.
In the present study, RAPD was used to investigate genetic polymorphisms in three related herbs, P. ginseng, P. quinquefolium, and P. notoginseng. Subsequently, species-specific fragments were recovered, cloned and sequenced. The sequence data were used to design unique primers to develop a specific PCR-based identification method for P. ginseng. We then verified this method using several medicinal herb samples. The method reported here enabled the authentication of P. ginseng from other crude drugs such as P. quinquefolium, P. notoginseng and other Chinese traditional medical herbs. Our method has several advantages: 1) it is simple and costs- effective, 2) the results enable accurate identification of P. ginseng that is rapid, reliable and meets the needs of the market.
2 Materials and methods
2.1 Plant materials and DNA isolation
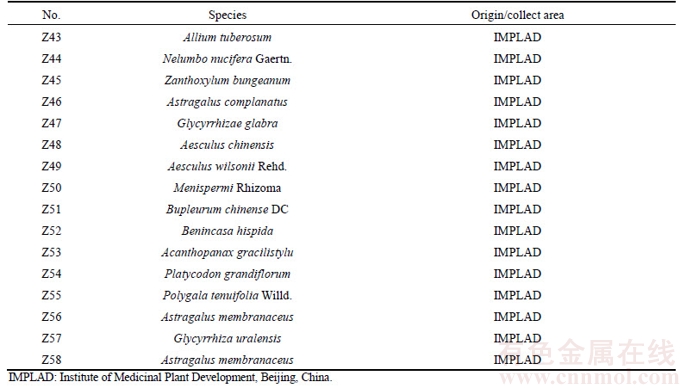
A total of 134 batches of samples were used in this study, including 33 batches of P. ginseng (Table 1), 14 batches of P. quinquefolium (Table 1), 19 batches of P. notoginseng (Table 1), 10 batches of adulterants (Table 2), and 58 batches of other traditional Chinese medicinal substances (Table 3). All samples were identified in the Hunan Institute for Drug Control and confirmed by our research group using DNA barcodes. Total genomic DNA was isolated using the CTAB method as described earlier [16].
2.2 RAPD analysis
A total of 120 RAPD primers were purchased from SBS Genetech Co. Ltd. (China). Two samples each of P. ginseng, P. quinquefolium and P. notoginseng were used for initial primer screening to identify primers that produce unique amplification bands. The identified primers were then confirmed by screening a larger number of samples. Each random primer was used for amplification at least twice. RAPD analysis was performed as previously described [15].
Table 1 Information about P. ginseng, P. quinquefolium, and P. notoginseng samples
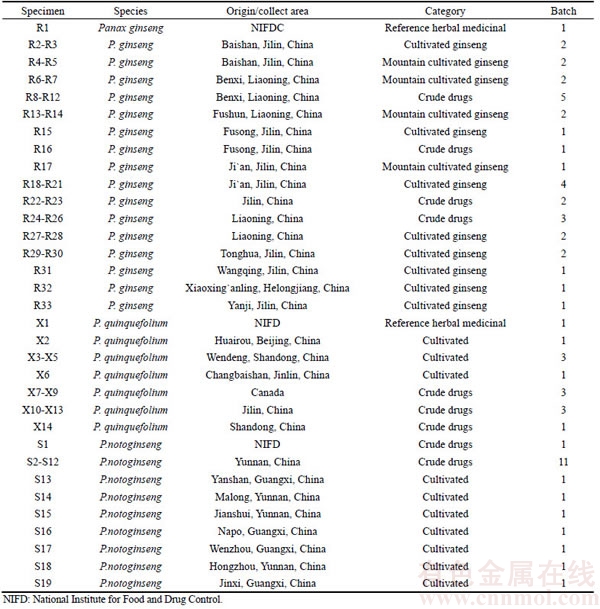
Table 2 Information about P. ginseng adulterant samples
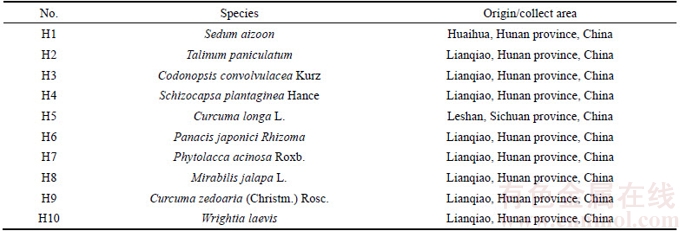
Table 3 Information about other common traditional Chinese medicinal herb samples used in study

Continued
2.3 Cloning, sequencing and designing SCAR primers
The RAPD band specific to P. ginseng was cloned into the pGM-T vector. The positive plasmids were bi-directionally sequenced by Biosune Biotech Co. Ltd. (China) using the universal primer pair T7/SP6. To ensure the accuracy of the sequences, at least two clones were sequenced [17, 18]. The sequences were assembled and edited using DNAStar 4.05 software (DNAStar, Inc.). The nucleotide sequence of the fragment C-12 was deposited in Genbank and primers to test P. ginseng specific to the identified sequences were designed using the primer-design software Oligo 6.
2.4 Development and optimization of PCR-based identification method for P. ginseng
The PCR reaction mixture consisted of 5.0 μL of 5× Primer STAR Buffer (Mg2+ plus), 2.5 μL of dNTPs (2.5 mmol/L), 0.75 μL of each of the designed primers: F1 and R1 (10 μmol/L), 0.25 μL of PrimeSTAR HS DNA polymerase (2.5 U/μL, Takara, China), and 1.0 μL of DNA template. The final volume of the reaction mixture was adjusted to 25 μL with sterilized ddH2O.
The amplification conditions were as follows: initial denaturation at 95°C for 3 min, 30 cycles of denaturation at 98°C for 10 s, annealing at 52°C for 15 s, extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min. The reactions were performed in a Veriti PCR thermal cycler (Applied Biosystems, USA).
This study also investigated the effect of different annealing temperatures, numbers of amplification cycles, amounts of DNA template, and types of Taq enzyme on the performance of the PCR-based identification method. Sterilized ddH2O was used as a blank control. Additionally, the universal primer pair ITS2 (ITS2F: ATGCGATACTTGGTGTGAAT; ITS3R: GACGCTTCTCCAGACTACAAT) was used to perform DNA amplification of all samples to be tested to verify the reliability of the template DNA.
2.5 Verification of P. ginseng-specific SCAR marker
DNA samples from 134 medicinal herb samples (Tables 1–3) were used as templates for P. ginseng-specific PCR amplification. The details of the reaction system, reaction conditions, and detection method were the same as those described in Section 2.4.
3 Results and discussion
3.1 Screening for P. ginseng-specific RAPD markers
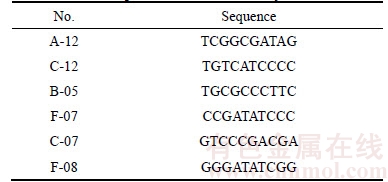
RAPD amplification assays performed using 120 random primers revealed that random primers A-12, B-05, C-07, C-12, F-07, and F-08 (Table 4) produced unique amplification products (Figure 1). Of these, random primers A-12 (Figure 1(a)) and B-05 (Figure 1(b)) produced unique bands in P. notoginseng, random primers C-07 (Figure 1(c)) and F-08 (Figure 1(d)) produced unique bands in P. quinquefolium, and random primers F-07(Figure 1(e)) and C-12 (Figure 1(f)) produced unique bands in P. ginseng. Figure 1(f) shows the results of RAPD amplification using the random primer C-12, with a P. ginseng specific RAPD marker C-12 at approximately 600 bp, and no markers for the P. quinquefolium and P. notoginseng samples, highlighting the specificity of the primers.
Table 4 Random primers for RAPD analysis
3.2 Conversion to SCAR markers
The six identified unique RAPD fragments were recovered from gels, purified and cloned into TA vectors. The clones obtained were sequenced using universal primers, and the corresponding nucleotide sequences were obtained. To confirm whether the RAPD marker C-12 is specific for P. ginseng, its nucleotide sequence was subjected to BlastN homology search against the nonredundant GenBank database. The search results showed no significant match with any previously determined sequences. The nucleotide sequence of the P. ginseng-specific marker C-12 has been submitted to GenBank with the accession number KU553472. Finally, an identification primer pair (F1: 5’CCCGACTCAAAATCGAAGT3’, R1: 5’AAGCAAGAAGCAAGGGTAACATA3’; Chinese Patent Application Number: 201510288554.0) was designed based on the obtained sequence information using Oligo 6 primer design software.
3.3 Development and optimization of SCAR identification method specific for P. ginseng
The primers F1 and R1 were subsequently used to develop the SCAR identification method specific for P. ginseng. As expected, the amplification of P. ginseng produced a band of approximately 300 bp in size, which also appeared in P. quinquefolius and P. notoginseng. Interestingly, a band at approximately 130 bp was observed only in P. ginseng but was absent among P. quinquefolius and P. notoginseng amplicons. These results suggested that this unique band pattern might distinguish P. ginseng from P. quinquefolius and P. notoginseng (Figure 2). To investigate the specificity and stability of this band pattern, it was necessary to study the effect of the annealing temperature, the number of cycles, the amount of template DNA, and the fidelity of the DNA polymerase used.
To determine the most suitable annealing temperature for amplification, various annealing temperatures (52, 54, 56 and 58°C) were tested. The results demonstrated that at an annealing temperature of 52–58°C, amplification of all P. ginseng samples generated two unique fragments at approximately 300 bp and 130 bp (Figure 2(a)). Considering the effects of annealing temperature on primer amplification efficiency and specificity, we selected 56°C as the ideal annealing temperature.
Various numbers of amplification cycles (30, 33, 36 and 39) were tested to determine the optimum number of amplification cycles. The results showed that 30 cycles were sufficient to produce a satisfactory intensity of bands(Figure 2(b)). To minimize the possibility of appearance of non-specific bands caused by excessive amplification cycles, 30 was selected as an ideal number of cycles for the PCR assay to ensure the accuracy of results.
To optimize the amount of DNA to be used as a template for the PCR assay, the DNA concentration was adjusted to approximately 30 ng/μL and various volumes of DNA (0.2, 1, 2 and 3 μL) were used in 25-μL PCR reactions. The results indicated that satisfactory PCR amplification was obtained with 0.2–3 μL of DNA template (equivalent to 6–90 ng of total DNA). It was observed that at sufficient number of cycles (30 cycles), the amplified bands became brighter with increasing amount of template DNA, with no effect on the results (Figure 2(c)).
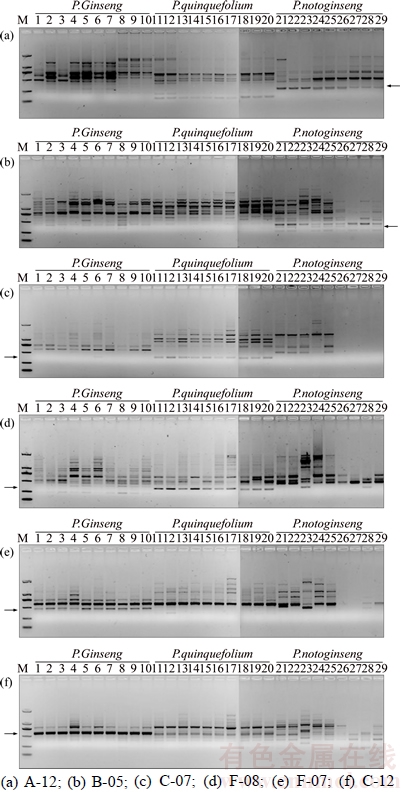
Figure 1 RAPD profiles of P. ginseng, P. quinquefolius and P. notoginseng amplified by primer: (Lanes 1 through 10 are P. Ginseng samples R2 through R11 in Table 1, respectively. Lanes 11 to 20 are P. quinquefolius samples X2 to X11 in Table 1, respectively. Lanes 21–29 are P. Notoginseng samples S2 to S10 in Table 1, respectively. M is the DNA marker, indicating DNA sizes of 2000 bp, 1000 bp, 750 bp, 500 bp, 250 bp, and 100 bp, from top to bottom. The arrow indicates the polymorphic band)
To establish the effect of DNA polymerases with different fidelities on amplification,high-fidelity HS DNA polymerase with regular buffer (Takara), high-fidelity HS DNA polymerase with GC buffer (Takara), high-fidelity PCR Master (Roche), Phanta Super-Fidelity DNA polymerase (Vazyme), and regular Taq enzyme (Tiangen) were used for PCR amplification in separate reactions. Specific amplification bands were observed when HS DNA polymerase (Takara) was used. The results also showed that with the same HS DNA polymerase, different buffers resulted in significantly different intensities of the amplified bands (Figure 2(d)). Overall, high-fidelity PrimeSTAR HS DNA polymerase with regular buffer (Takara) demonstrated the best performance.
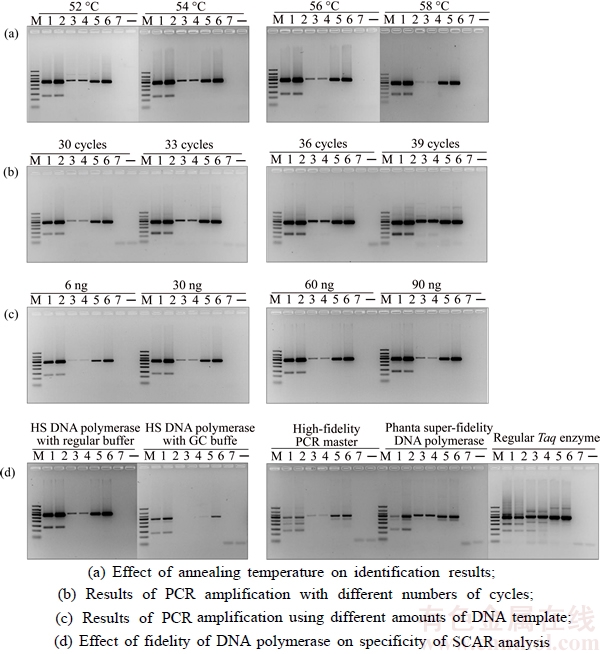
Figure 2 Optimization of PCR-based identification method for P. Ginseng:(Lanes 1 and 2 are sample R2 and R6 of P. ginseng (Table 1), respectively. Lanes 3 and 4 are sample X2 and X3, P. quinquefolium (Table 1), respectively. Lanes 5 and 6 are Sample S2 and S3 of P. notoginseng (Table 1), respectively. Lane 7 is adulterants Mirabilis jalapa L. Lane 8 is the negative control. M is the DNA 50 bp marker (Tiangen), indicating DNA sizes of 500 bp, 400 bp, 350 bp, 300 bp, 250 bp, 200 bp, 150 bp, 100 bp, and 50 bp, from top to bottom)
3.4 Verification of SCAR markers
The optimized PCR-based identification system for P. ginseng was further evaluated by testing the 134 batches of samples. In this study, specific PCR products were produced from these samples (Tables 1–3) after amplification using ITS2 universal primers, suggesting that the quality of DNA from the samples met the assay requirements. The electropherogram (Figure 3(a)) revealed that amplification of all P. ginseng samples produced two unique bands at approximately 300 bp and 130 bp. As expected, this band pattern was absent in P. quinquefolium (Figure 3(b)), P. notoginseng (Figure 3(c)), Sedum aizoon, Talinum paniculatum, Codonopsis convolvulacea Kurz, Schizocapsa plantaginea Hance, Curcuma longa L., Panacis Japonici Rhizoma, Phytolacca acinosa Roxb. (Figure 3(c)), and 58 other traditional Chinese medicinal substances studied (Figure 3(e)). These results indicated that the identified ginseng-specific SCAR marker could be used to specifically distinguish P. ginseng from other medicinal herbs.
Figure 3 SCAR analysis of P. ginseng and their adulterants amplified by specific primers F1 and R1
4 Conclusions
In this study, we successfully developed a SCAR assay to identify P. ginseng among traditional Chinese medicinal products. Our achievements can be summarized as follows:
1) Random primers A-12, B-05, C-07, C-12, F-07, and F-08 produced unique amplification products in P. ginseng, P. quinquefolium, and P. notoginseng in RAPD amplification assays.
2) The RAPD marker was converted to a stable SCAR marker, by designing a P. ginseng-specific identification primer pair (F1: 5’CCCGACTCAAAATCGAAGT3’, R1: 5’AAGCAAGAAGCAAGGGTAACATA3’; Chinese Patent Application Number: 201510288554.0).
3) The SCAR marker-based identification system for Panax ginseng was developed, after optimization of the reaction conditions, which generated a unique band pattern (two DNA fragments at approximately 300 bp and 130 bp), only in P. ginseng samples. This pattern was not observed upon amplification of P. quinquefolium, P. notoginseng, Sedum aizoon, Talinum paniculatum, Codonopsis convolvulacea Kurz, Schizocapsa plantaginea Hance, Curcuma longa L., Panacis Japonici Rhizoma, Phytolacca acinosa Roxb, and 58 other medicinal herb samples examined in this study. The observed results confirmed that the proposed method could be used to authenticate P. ginseng. The proposed method adopts a PCR-based specific SCAR marker technology to identify ginseng and its decoction products and involves a series of simple processes of DNA extraction, PCR amplification, and electrophoresis. Owing to advantages such as easy operation, high specificity, and requirement of a small sample size, this method is a practical and feasible tool for P. ginseng identification, with many potential applications. The SCAR analysis is expected to be particularly relevant for the establishment of reliable DNA fingerprinting for quality control of P. ginseng.
References
[1] ATTELE A S, WU Ji-an, YUAN Chun-su. Ginseng pharmacology: Multiple constituents and multiple actions [J]. Biochemical Pharmacology, 1999, 58(11): 1685–1693.
[2] CHANG Y S, SEO E K, GYLLENHAAL C, BLOCK K I. Panax ginseng: a role in cancer therapy? [J]. Integrative Cancer Therapies, 2003, 2(1): 13–33.
[3] CHENG Yong, SHEN Li-hong, ZHANG Jun-tian. Anti-amnestic and anti-aging effects of ginsenoside Rg1 and Rb1 and its mechanism of action [J]. Acta Pharmacologica Sinica, 2005, 26(2): 143–149.
[4] CHOO M K, PARK E K, HAN M J, KIM D H. Antiallergic activity of ginseng and its ginsenosides [J]. Planta Medica, 2003, 69(6): 518–522.
[5] SHIN H R, KIM J Y, YUN T K, MORGAN G, VAINIO H. The cancer-preventive potential of Panax ginseng: A review of human and experimental evidence [J]. Cancer Causes & Control, 2000, 11(6): 565–576.
[6] WANG Hong-wei, PENG Da-cheng, XIE Jing-tian. Ginseng leaf-stem: Bioactive constituents and pharmacological functions [J]. Chinese Medicine, 2009, 4(1): 20. DOI: https://doi.org/10.1186/1749-8546-4-20.
[7] HE Yan-qing. The discuss ginseng market situation and the authenticity of identification [J]. China Practical Medical, 2011, 6(18): 226–227.
[8] WU Xue-ling, LIU Li-li, ZHANG Zhen-zhen, DENG Fan-fan, LIU Xin-xing. Molecular characterization of Acidithiobacillus ferrooxidans strains isolated from different environments by three PCR-based methods [J]. Journal of Central South University, 2015, 22(4): 455-465.
[9] CHEN Li-jing, QI Xin, WANG Yu-kun, ZHANG Li, GUO Zhi-fu, LIN Jing-wei, SONG Yu-ning, ZHONG Ming. Identification of Schisandra sphenanthera and S. chinensis by random amplified polymorphic DNA sequence characterized applied region] [J]. China Journal of Chinese Materia Medica, 2011, 36(22): 3083–3085.
[10] CUI Zhan-hu, LONG Ping, WANG Ying-li, BAI Xiao-rong, YUAN Yuan, LI Min-hui. Application and prospect of DNA molecular markers in the identification of Chinese Medicine [J]. Journal of Chinese Medicinal Materials, 2015, 38(1): 188–192.
[11] KIRAN U, KHAN S, MIRZA K J, RAM M, ABDIN M Z. SCAR markers: A potential tool for authentication of herbal drugs [J]. Fitoterapia, 2010, 81(8): 969–976.
[12] SUN Tao, TENG Shao-na, KONG De-ying, SONG Yun, XU Jin, LI Ying-guo, WANG Yu, LI Ming-fu. DNA barcoding used in the identification of ginseng [J]. China Biotechnology, 2013, 33(4): 143–148.
[13] ZHOU Tao, XIE Yu, ZHANG Li-yan, WEI Sheng-hua, JIN Yan-lei. Study on sequence characterized amplified region (SCAR) markers of polygonum capitatum [J]. China Journal of Chinese Materia Medica, 2013, 38(16): 2577–2580.
[14] Tanaka H, Fukuda N, Shoyama Y. Identification and differentiation of Panax species using ELISA, RAPD and eastern blotting [J]. Phytochemical Analysis, 2006, 17(1): 46–55.
[15] Jung Juyeon, Kim Kyung-hee, Yang Kiwoung, Bang Kyong-hwan, Yang Tae-Jin. Practical application of DNA markers for high-throughput authentication of Panax ginseng and Panax quinquefolius from commercial ginseng products [J]. Journal of Ginseng Research, 2014, 38(2): 123–129.
[16] PIRTTIL A M, HIRSIKORPI, M, K
A M, HIRSIKORPI, M, K M
M R
R INEN T, JAAKOLA L, HOHTOLA A. DNA isolation methods for medicinal and aromatic plants [J]. Plant Molecular Biology Reporter, 2001, 19(3): 273–273.
INEN T, JAAKOLA L, HOHTOLA A. DNA isolation methods for medicinal and aromatic plants [J]. Plant Molecular Biology Reporter, 2001, 19(3): 273–273.
[17] SOLIERI L, GIUDICI P. Development of a sequence- characterized amplified region marker-targeted quantitative PCR assay for strain-specific detection of Oenococcus oeni during wine malolactic fermentation [J]. Applied and Environmental Microbiology, 2010, 76(23): 7765–7774.
[18] XIA Jin-lan, ZHANG Qian, ZHANG Rui-yong, PENG Juan-hua, PENG An-an, ZHAO Xiao-juan, NIE Zhen-yuan, QIU Guan-zhou. Isolation and characterization of acidophilic bacterium from Dongxiangshan Mine in Xinjiang Province, China [J]. Journal of Central South University, 2010, 17(1): 50-55.
(Edited by YANG Hua)
中文导读
用于药品检验的人参SCAR标记研究
摘要:为了研究人参、西洋参和三七的基因多态性,使用120条随机引物进行了随机扩增多态性DNA分析。将筛选获得的人参特异性RAPD标记C-12进行T-A克隆、测序(GenBank登录号KU553472)。根据序列分析结果,设计了一对特异性引物,经优化反应条件,建立了人参的SCAR标记鉴别体系。在所有的33批人参标本中,均能稳定地获得约300 bp和130 bp的2条阳性扩增带;在混伪品以及西洋参、三七和其他中药材共计101批标本中均为阴性扩增。因此,建立了一种快速、有效的能准确、特异性地鉴别人参的PCR方法,可用于人参的种质资源保护、利用及其基原鉴定。
关键词:人参;随机扩增多态性DNA(RAPD);特征序列扩增标记(SCAR);分子鉴定
Foundation item: Project(2014ZX09304307-002) supported by the Major Program of Science and Technology Foundation of China; Project supported by Technology Platform for Quality/Safety Inspection and Risk Management of Traditional Chinese Medicine, China; Project(2014SK2001) supported by the Key Program Foundation of Hunan Provincial Science & Technology Department, China; Project(XSYK-R201502) supported by the Hunan Provincial Food and Drug Administration under Key Project of Science and Technology for Food and Drug Safety, China
Received date: 2017-04-26; Accepted date: 2017-10-30
Corresponding author: LI Wen-li, Professor; Tel: +86–731–82275866; E-mail: 1838675867@qq.com; LI Wen-zhang, Professor; Tel: +86–13874992656; E-mail: liwenzhang@csu.edu.cn; ORCID: 0000-0002-1020-0554