
Screen biomarkers of human lung squamous carcinoma by serological proteome analysis of HTB-182
LI Cui(李 萃), LI Xin-ying(李新营), TANG Can-e(汤参娥), YI Hong(易 红), DUAN Chao-jun(段朝军)
Key Laboratory of Cancer Proteomics of Chinese Ministry of Health, Xiangya Hospital,
Central South University, Changsha 410008, China
Received 10 April 2006; accepted 25 April 2006
Abstract:
Carcinogenesis of lung squamous carcinoma is a complex process involving multiple events and steps. At present, there are very few special lung cancer molecular markers for an “early-stage” diagnosis and prognosis evaluation. To identify tumor-associated antigens, serological proteome analysis (SERPA) of human lung squamous carcinoma cell line HTB-182 are performed. We characterized sixteen differentially expressed proteins which react with lung squamous carcinoma patient sera while not react with control sera. Some of these candidate lung squamous carcinoma–associated antigens were metabolic enzymes, such as triosephosphatase isomerase (TPIS). Some proteins were involved in the regulation of cell cycle and signal transduction. The results will provide scientific foundation for screening the molecular biomarkers used to diagnose and treat lung squamous carcinoma, as well as to elevate the patient’s prognosis and provide new clue for the research of lung squamous carcinogenic mechanism. Thus, SERPA represents a valuable approach for the identification of differentially expressed proteins, which might be used as markers for the diagnosis and prognosis of lung squamous carcinoma.
Key words:
biomarker; HTB-182; SERPA; 2-DE; MALDI-TOF-MS;
1 Introduction
Lung cancer is a challenging worldwide clinical problem, and its overall incidence is increasing. According to histological diagnosis, lung cancer comprises a broad spectrum of tumors that include squamous carcinoma, adenocarcinoma, large-cell carcinoma, and small-cell carcinoma. Squamous carcinoma is the most common type, accounting for 30%-50% in all of lung cancer patients in China[1]. Despite a number of efforts are currently performed to characterize lung cancer using molecular biological, cytogenetic, immunohistochemical as well as proteome-based techniques, corresponding improvements in outcome are not yet apparent. Recently, a combination of two-dimensional polyacrylamide gel electrophoresis (2-DE) expression profiling of tumor cell lines or tissues and immunoblotting with sera from patient and healthy people (control) termed SERPA might serve as a powerful tool for the identification of tumor associated antigen (TAA) because that pathological changes within an organ might be reflected in proteomic patterns in serum. These antigens eliciting a humoral response might be employed in cancer screening, diagnosis or in establishing prognosis as well as novel therapeutic targets. SERPA, a proteomic-based approach using western blotting experiments with sera from cancer patients, followed by the identification of the target proteins by mass spectrometry and database searching, has permitted the screening of autoantibodies of patients with cancer. So far, SERPA has been successfully applied to different types of cancer such as renal cellular carcinoma, neuroblastoma, lung adenocarcinoma, breast cancer or hepatocellular, several useful autoantigens have been identified[2-10]. Our study was engaged in identification of new biological markers of human lung squamous carcinogenesis by employing this approach.
2 Experimental
2.1 Cell lines and sera preparation
Human lung squamous carcinoma cell line HTB-182 were propagated in RPMI 1 640 medium supplemented with 10% fetal bovine serum in humidified 5%CO2 atmosphere at 37 ℃. HTB-182 was seeded in 100 mm dishes, After being washed twice with ice cold phosphate-buffered saline, cells were collected with a scraper and cetrifuged at 4 ℃, 2 000 r/min for 10 min×2. The cell pellets(approaximately 1×107 cell) were lysed in 200 μL lysis buffer consisting of 7 mol/L urea, 2 mol/L thiourea, 100 mmol/L DTT,5 mmol/L PMSF,4% CHAPS,40 mmol/L Tris,2% pharmalyte,1 mg/mL DNase I, and 0.25 mg/mL RNase A; votexed, incubated (room temperature, 2 h). The mixture was centrifuged (15 000 r/min, 30 min, 4 ℃)[11]. The supernatant was the total protein solution. The concentration of the total proteins was assayed with the protein Assay Kit (Amersham Biosciences).
Tumor sera were obtained from patients with lung squamous cancer after informed consent. All samples were diagnosed by histopathology. Patients, medical records were reviewed. Additionally, the control sera were attained from ten healthy volunteers. All serum samples were isolated from venous blood after informed consent. Sera were stored in liquid nitrogen.
2.2 IPG-2D PAGE
Samples were then loaded onto linear immobilized pH gradient(IPG) strips (pH 3-10 L, 130 mm×3 mm×0.5 mm, Amersham Biosciences) using 200 μg protein. Three gels were run simultaneously with equal amounts of the same protein. IPG-2D PAGE consisted of IEF that was performed using on IPGphor isoelectric focusing cell and Second-dimension SDS-PAGE (Amersham Biosciences)with using Ettan Dalt II system as described by manufacturer and GORG et al[12]. After separation, one (replica gel) was visualized by silver-based staining technique with the protein silver stain kit (Amersham Biosciences). The others were used for western blotting.
2.3 2-D Western blotting
Proteins were transferred to (NC) blotting membranes (Immobilon P, Millipore) using a semi-dry system(TE70 series Semi-Dry Transfer Unit, Amersham Biosciences) in transfer buffer (39 mmol/L glycine, 48 mmol/L Tris, 0.0375% SDS, 20% methanol) at 0.8 mA/cm2 for 70 min. The transfer efficiency was checked by staining of the membranes with Ponceau S. The membranes were blocked with 5% skimmed milk in TBS–T(2 h, room temperature). The membranes were incubated overnight at 22 ℃ with serum obtained either from patients or from controls(1∶150 dilution), washed and incubated with horseradish peroxidase -conjugated anti-human IgG antibody (1∶2 000 dilution) (Amersham Biosciences) for 1 h at room temperature. Immunodetection was accomplished by enhanced chemiluminescence (ECL; Amersham Biosciences) followed by autoradiography on hyperfilm MP (Amersham Biosciences). The imaging films exposed to the blots were densitometrically scanned using ImageScanner and matching was done by comparing the films of blots with the Ponceau S-stained membrane image and later with the replica gel.
2.4 Image analysis
The stained 2-DE gels, the Ponceau S-stained nitrocellulose membrane and the paired ECL-developed imaging films were scanned with LabScan software on Imagescanner (Amersham Biosciences) with 300 resolutions. The spot-intensity calibration, spot detection, background abstraction, matching, 1-D calibration, 2-D calibration were performed with ImageMaster 2D Elite 4.01 analysis software (Amersham Biosciences). Protein staining and immunological recognition of individual spots were quantified by calculation of spot volume after normalization of the image using the total spot volume normalization method multiplied by the total area of all the spots. The Ponceau S spots chosen as landmarks onto the image of the ECL film.
2.5 MALDI-TOF-MS & database analysis
Differential protein spots were excised from preparative gels using biopsy punches and transferred to a 1.5 mL siliconized Eppendorf tube. Proteins were in-gel digested as previously described[11]. The samples were analyzed with Applied Biosystems Voyager System 4307 MALDI-TOF(matrix-assisted laser desorption/ionization time of flight) Mass Spectrometer (ABI). The parameters were set up as follows: positive ion-reflector mode, accelerating voltage 20 kV, grid voltage 64.5%, mirror voltage ratio 1.12, N2 laser wavelength 337 nm, pulse width 3 ns, the number of laser shots 50, acquisition mass range 1 000-3 000 u, and delay 100 ns, and vacuum degree 5.332 88×10-5 Pa. A trypsin-fragment peak served as internal standard for mass calibration. A list of the corrected mass peaks was the peptide mass fingerprinting (PMF). Proteins were identified with PMF data by searching software Mascot (http://www.matrixscience.com) with using Mascot Distiller which can detect peaks by attempting to fit an ideal isotopic distribution to the experimental data. The searching parameters were set up as follows: the mass tolerance was ±0.5 u; the number of missed cleavage sites was allowed up to 1; the cysteine residue was modified as carbamidomethyl-cys; variable modifica-tions was oxidation (M); the minimum number of matched-peptides was 4; species was selected as HOMO SAPIENS (HUMAN); the peptide ion was [M+H]+; mass values were monoisotopic; the isotope masses were used; protein scores greater than 62 are significant (p<0.05).
3 Results and discussion
Serologic Proteome Analysis (SERPA) of human lung squamous carcinoma cell line HTB-182 was performed. The following strategy was pursued for the identification of proteins reacting with high-titer IgG molecules in lung cancer patient sera: three gels were run simultaneously with equal amounts of the same sample. One is silver stained as shown in Fig.1(a) with a total of 1 135±31 spots, the remaining gels are electro-blotted onto nitrocellulose (NC) membranes, then the NC membranes are stained with Ponceau S (Fig.1(b)). One blot is probed with autologous patient serum(shown in Fig.1(c)), the other with a control serum is obtained from a healthy donor(shown in Fig.1(d)). In order to measure the reproducibility, western blot imaging films of HTB-182 reacting with autologous patient sera and with control sera from the same patient is repeated for three times, respectively. The image analysis shows these western blot imaging films are reproducible. The differential spots are matched with an average matching rate of 95.3%. Well-resolved, reproducible 2-DE western blot imaging films of HTB-182 reacting with autologous patient sera and with control sera are obtained. In general, a greater number of reactive proteins were detected with sera from patients with lung cancer than with control sera.
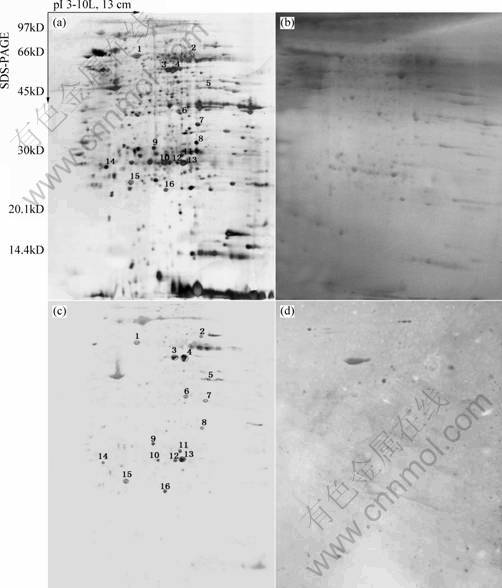
Fig.1 Two dimensional electrophoretic map and 2-DE Western blot imaging films of HTB-182 (Tagged spots represent identified proteins which react with lung squamous carcinoma patients autologous sera while not react with control sera): (a) Two dimensional electrophoretic map of HTB-182; (b) Image of ponceau S-stained nitrocellulose membrane; (c) 2-DE western blot imaging films of HTB-182 reacting with autologous patient sera; (c)2-DE western blot imaging films of HTB-182 reacting with control sera
As expected, Fig.1(c) shows more protein spots which apparently react with serum IgG than control Fig.1(d) which also shows few similar numbers of reactive spots. The results of this analysis are listed in Table 1.
In this study, the western-blotting imaging films reacting with autologous patient serum and with control serum were compared by using ImageMaster 2D Elite 4.01 analysis software, in which average 28±3 differentially expressed protein-spots were detected. The corresponding position of these differential spots in the silver replica gel was found by matching and comparing the films of blots with the Ponceau S-stained membrane image and later with the replica gel. Thereinto 22 spots which have the same reaction with lung squamous carcinoma patient sera while not with control sera in three times experiments were regarded as lung squamous cancer associated-antigen(Fig.1).
Twenty two differential protein-spots which reacted reproducibly with the autologous serum but not with the control serum were identified with MALDI-TOF-MS. These differential protein-spots were excised from the silver stained gels, and digested in-gel with trypsin. The peptide mass fingerprinting (PMF) maps were obtained by MALDI-TOF-MS, and calibrated with Trypsin auto-degraded peak (m/z=1 993.977 2). Fig.2 shows the PMF maps of the protein-spots W12. The PMF data were used to search the SWISS-PROT and NCBI databases with Mascot software. The resulting protein was determined by comprehensively considering the corresponding experimental pI, Mr, the number of matched-peptides, and the sequence coverage (Table 1). The score of sixteen protein exceed 62 in Mascot searching within twenty two differential protein-spots, the result can be considered credibility(p<0.05)(Fig.3). The data on all identified protein spots are described in Table 1.
Mass spectrometry is emerging as an important tool in biochemical research. In the past, mass spectrometry was confined to the realm of small molecules, large molecules did not survive the desorption and ionization process intact. More recently, the development of new “mild” desorption and ionization methods has revolutionized the analysis of large biomolecules, making mass spectrometry an important analytical tool
Table 1 Protein spots searched by Mascot software in database
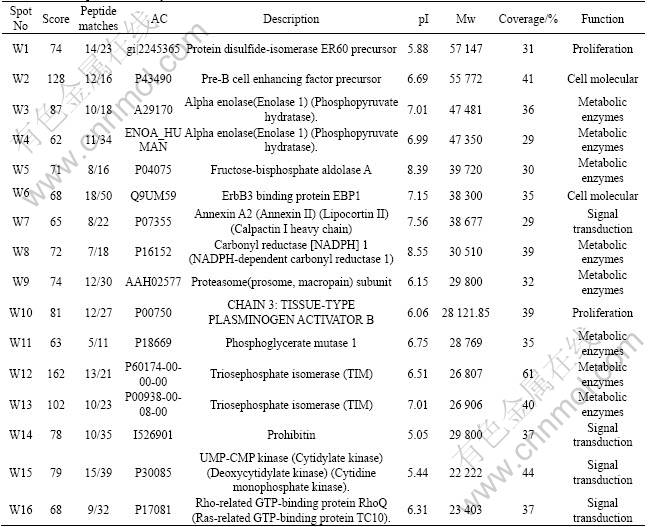
Fig.2 Peptide mass fingerprinting of protein spot W12 in HTB-182 2-DE map scanned by Mascot Distiller
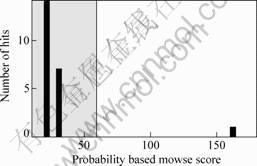
Fig.3 Score of peptide mass fingerprinting of W12 searched by Mascot (162)
for biological research[13]. With these new ionization technologies, mass spectrometry is applied into the realm of biomolecules, including screening the biomarker of tumor. Our previous studies based on 2-DE/MALDI-
TOF-MS have compared proteome of human lung squamous carcinoma and normal bronchial epithelial tissues and have identified differentially expressed proteins[11]. In order to further elucidate the carcinogenesis of lung squamous carcinoma, the SERAP proteomic study based on 2-DE western blot/MALDI-TOF-MS was performed in human lung squamous carcinoma cell lines HTB-182. The results show an IgG immunoreactivity against 16 proteins, including metabolic enzymes, cell molecular, and some proteins which are involved in the regulation of cell cycle and signal transduction. Among 16 proteins, eight metabolic enzymes such as protein disulfide-isomerase ER60 precursor, Alpha enolase and its isoform, Triosephosphate isomerase and its isoform, Fructose- bisphosphate aldolase A, Carbonyl reductase1 and Phosphoglycerate mutase 1 exhibited differential reaction between lung squamous carcinoma patient sera and health control sera. These data show that some metabolic enzymes differentially react with patient’s autologous sera. Thus it can be seen, the lung squamous carcinoma is often associated with a number of metabolic and biochemical alterations. The other proteins such as Annexin Ⅱ is a calcium- and phospholipids-binding protein and a substrate for protein-tyrosine kinases. Recently, Annexin Ⅱ heterotetramer has been shown to play an important role in degradation of extracellular matrix proteins, an important step for tumor cell local invasion, angiogenesis, and metastasis. It is assumed that Annexin Ⅱmay play an important role in the carcinogenesis of carcinoma[14]. Rho-related GTP-binding protein RhoQ plays pivotal roles in actin cytoskeletal organization, Golgi vesicular trafficking, receptor endocytosis, and cell cycle progression[15]. These results suggest that carcinogenesis of lung squamous carcinoma is a complex process involving in some proteins which are cell cycle regulator, signal transduction and immune modulation molecular and so on.
Sixteen proteins, which are lung squamous carcinoma–associated antigens, were identified by PMF. Ten of them such as protein disulfide-isomerase ER60 precursor, Alpha enolase and its isoform, Triosephosphate isomerase and its isoform, Fructose-bisphosphate aldolase A, Carbonyl reductase1, Phosphoglycerate mutase 1, Pre-B cell enhancing factor precursor and Annexin Ⅱ were also identified in the SERPA study of lung squamous carcinoma tissues, which validates the reliability of the identified results and further supports that these lung squamous carcinoma-associated antigens may be the candidate molecular biomarkers for the diagnosis and therapy of lung squamous carcinoma and also suggest an important complementary role for proteomics in identification of molecular abnormalities important in cancer development and progression. The function and acting mechanism of these proteins await further study.
4 Conclusions
The SERAP proteomic investigation based on 2-DE western blot/MALDI-TOF-MS is an effective platform to study the human lung squamous carcinoma. The results presented will benefit identification of potential tumor markers and understanding of the mechanisms of human lung squamous carcinoma.
References[1] LOPEZ A D. Counting the dead in China: measuring tobacco’s impact in the developing world[J]. Br Med J, 1998, 317: 1399.
[2] LE NAOUR F. Contribution of proteomics to tumor immunology[J]. Proteomics, 2001, 1(10): 1295-1300.
[3] SELIGER B, KELLNER R. Design of proteome-based studies in combination with serology for the identification of biomarkers and novel targets[J]. Proteomics, 2002, 2(12): 1645-1651.
[4] STOCKERT E, JAGER E, CHEN Y T, SCANLAN M J, GOUT I, KARBACH J, ARAND M, KNUTH A, OLD L J. A survey of humoral immune response of cancer patients to a panel of human tumor antigens[J]. J Exp Med, 1998, 187:1349-1354.
[5] NAOUR F L, BRICHORY F, BERETTA L, HANASH S M. Identification of Tumor-Associated antigen using proteomics[J]. Technology In Cancer Research and Treatment, 2002, 1(4): 257-262.
[6] NAOUR F L, BRICHORY F, MISEK D E, BRECHOT C, HANASH S M, BERETTA L. A distinct repertoire of autoantibodies in hepatocellular carcinoma identified by proteomic analysis[J]. Molecular & Cellular Proteomics, 2002, 1(3): 197-203.
[7] KELLNER S, LICHTENFELS R, ATKINS D, BUKUR J, ACKERMANN A, BECK J, BRENNER W, MELCHIOR S, SELIGER B. Targeting of tumor associated antigens in renal cell carcinoma using proteome-based analysis and their clinical significance[J]. Proteomics, 2002, 2(12): 1743-1751.
[8] LE NAOUR F, MISEK D E, KRAUSE M C, DENEUX L, GIORDANO T J, SCHOLL S, HANASH S M. Proteomics-based identification of RS/DJ-1 as a novel circulating tumor antigen in breast cancer[J]. Clin Cancer Res, 2001, 7(11): 3328-3335.
[9] PRASANNAN L, MISEK D E, HINDERER R, MICHON J, GEIGER J D, HANASH S M. Identification of beta-tubulin isoforms as tumor antigens in neuroblastoma[J]. Clin Cancer Res, 2000, 6(10): 3439-3456.
[10] BRICHORY F, BEER D, LE NAOUR F, GIORDANO T, HANASH S. Proteomics-based identification of protein gene product 9.5 as a tumor antigen that induces ahumoral immune response in lung cancer[J]. Cancer Res, 2001, 61(21): 7908-7912.
[11] LI C, XIAO Z Q, CHEN Z C, ZHANG X P, LI J L, WU X Y, LI X Y. Proteome analysis of human lung squamous carcinoma[J]. Proteomics, 2006, 6(2): 547-558.
[12] GORG A, OBERMAIER C, BOGUTH G, HARDER A, SCHEIBE B, WILDGRUBER R, WEISS W. The current state of two-dimensional electrophoresis with immobilized pH gradients[J]. Electrophoresis, 2000, 21: 1037-1053.
[13] SIUZDARK G. Mass Spectrometry for Biotechnology[M]. California: Academic Press, 1996.
[14] DANIELSEN E M, VAN DEURS B, HANSEN G H. “Nonclassical” secretion of annexin A2 to the lumenal side of the enterocyte brush border membrane[J]. Biochemistry, 2003, 42(49): 14670-14676.
[15] LIN Q, FUJI R N, YANG W, CERIONE R A. RhoGDI is required for Cdc42-mediated cellular transformation[J]. Curr Biol, 2003, 13(17): 1469-1479.
Foundation item: Project (2001CB5102) supported by the National Basic Research Program of China; Project (30500558) supported by the National Natural Science Foundation of China
Corresponding author: DUAN Chao-jun; Tel: +86-731-4327298; Fax: +86-731-4327332; E-mail: Duancj2001@yahoo.com.cn


