
Sulfur activation-related extracellular proteins of Acidithiobacillus ferrooxidans
ZHANG Cheng-gui(张成桂), ZHANG Rui-yong(张瑞永), XIA Jin-lan(夏金兰),
ZHANG Qian(张 倩), NIE Zhen-yuan(聂珍媛)
Key Laboratory of Biometallurgy of Ministry of Education, School of Minerals Processing and Bioengineering,
Central South University, Changsha 410083, China
Received 20 September 2008; accepted 5 November 2008
Abstract:
The fractions of the extracellular proteins of Acidithiobacillus ferrooxidans grown on two different energy substrates, elemental sulfur and ferrous sulfate, were selectively prepared with hot water treatment and distinctly shown by two-dimensional gel electrophoresis. Some protein spots with apparently higher abundance in sulfur energy substrate than in ferrous sulfate energy substrate were identified by using MALDI-TOF/TOF. Based on peptide mass fingerprints and bioinformatical analysis, the extracellular proteins were classified according to their functions as conjugal transfer protein, pilin, vacJ lipoprotein, polysaccharide deacetylase family protein, Ser/Thr protein phosphatase family protein and hypothetical proteins. Several extracellular proteins were found abundant in thiol groups and with CXXC functional motif, these proteins may be directly involved in the sulfur activation by use of their thiol group (Pr-SH) to bond the elemental sulfur.
Key words:
sulfur oxidation; extracellular proteins; Acidithiobacillus ferrooxidans;
1 Introduction
Elemental sulfur as the intermediate product of the bioleaching process may be accumulated and form a sulfur layer on the mineral surface, leading to the modification of the electrochemical properties of mineral surface and the inhibition of the bioleaching process [1-2]. It is apparently significant to elucidate and find the efficient way to eliminante this sulfur layer. Actually, the elemental sulfur in bioleaching is basically chemically inert, and it can exclusively be eliminated by bio-oxidation of acidophilic sulfur-oxidizing bacteria in typical acidic environments[1,3].
It is known that oxidation of elemental sulfur by acidophilic sulfur-oxidizing bacteria is a complex process that involves principally the attachment of cells to sulfur particles, the transportation of sulfur through the outer membrane and the oxidation of sulfur in the periplasmic space[4-5]. A prerequisite for initial attachment of bacterial cells to the sulfur particles is that the extracellular polymeric substances (EPS) excreted by cells mediate this contact, by modifying the surfaces of both the sulfur particles and the cells themselves[6-7]. EPS bridge the contact between sulfur and cells to overcome the sulfur surface hydrophobicity, and assure cells to adsorb onto sulfur surface. The extracellular proteins and outer-membrane proteins were considered playing the key role in the sulfur mobilization and sulfur transportation, respectively[8-9]. Acidithiobacillus thiooxidans grown on elemental sulfur exposed extensive membranous blebbing of outer membrane which coated the surface of elemental sulfur particles to promote the dissolution of sulfur to colloidal sulfur[3]. A sulfur binding surface protein of 40 ku has been identified in A. ferrooxidans grown on elemental sulfur, which locates on the flagella, has thiol groups and adheres to elemental sulfur particles[10], and several outer-membrane proteins have been suggested to be involved in the adhesion of A. ferrooxidans to various surfaces and highly regulated by different energy substance[10-12]. However, up to now, few studies have been devoted to evaluating the extracellular proteins for adaptation of bacterial cells to the hydrophobic sulfur and activation/mobilization of sulfur by A. ferrooxidans cells.
The present work was dedicated to analyzing the changes of A. ferrooxidans extracellular proteins when the bacteria cells are cultured in two different energy substrates, elemental sulfur and ferrous sulfate, in order to further understand the sulfur mobilization process.
2 Experimental
2.1 Bacterial strains and cultivation conditions
A. ferrooxidans ATCC23270 was obtained from the American Type Culture Collection. It was cultivated at 30 ℃ in 9K medium containing 5.0 g/L of S0 or 22.5 g/L ferrous sulfate and the following components (g/L): (NH4)2SO4, 3.0; MgSO4·7H2O, 0.5; KCl, 0.1; K2HPO4, 0.5 and Ca (NO3)2, 0.01. The initial pH of these media was adjusted to 2.0 with KOH.
2.2 Isolation and partial purification of extracellular proteins
Bacteria cells were harvested in the exponential growth phase by centrifugation from an initial 8 L sample of bacterial culture. The pellet was washed with distilled water (pH=2), and then resuspended in 10 mL of distilled water (pH=2) at 60 ℃ for 40 min in a shaking water bath to shed the extracellular proteins from cells. The supernatants obtained on centrifugation after all treatments were filtered through 0.22 μm cellulose acetate filters in a Pyrex filtering apparatus (Millipore) to ensure that samples were free of cells. The supernatants with extracellular proteins were extensively dialyzed against distilled water at 4 ℃ in sterilized dialysis bag (cutoff, 3 500 Da). The remaining solution of crude extracellular proteins was freeze-dried and stored at -80 ℃.
2.3 Two-dimensional gel electrophoresis (2-DE)
2-DE was performed with the IPGphor system and Ettan DALT II system (GE). After electrophoresis, proteins were visualized by Coomassie brilliant blue R-250 staining.
2.4 Image acquisition and analysis
All experiments were duplicated and the corresponding gels were realized in triplicate. Two- dimensional gels were scanned using a Uniscan scanner (Tsinghua Unisplendour). The number of protein spots on the 2-DE gels was analyzed using Imagemaster software version 7.2 (GE).
2.5 In-gel tryptic digestion of 2-DE spots and MALDI-TOF/TOF
Protein spots were excised and subjected to in-gel digestion. Peptide mass fingerprints were obtained by using an Autoflex MALDI-TOF/TOF trandem mass spectrometer (Bruker Daltonik Bremen), with delayed extraction and reflection[13-14].
2.6 Database searches for protein identification and protein sequence analysis
Peptide mass fingerprintings were analyzed by using MASCOT search engine (www.matrixscience.com) based on SwissProt and AFE-3217 protein databases. The functional categories for the different identified proteins were obtained from the genomic sequence of A. ferrooxidans (www.tigr.org). The presence of export signals for each protein found in the periplasmic fraction was predicted by using SignalP (www.cbs.dtu.dk/ services/signalP), TatP (www.cbs.dtu.dk/services/TatP- 1.0), SecretomeP (www.cbs.dtu.dk/services/SecretomeP), and LipoP(www.cbs.dtu.dk/services/LipoP). The predicted subcellular locations of the proteins coded by the different ORFs analyzed were obtained using PENCE proteome analyst (http://pasub.cs.ualberta.ca:8080/pa/ Subcellular) and CELLO (cello.life.nctu.edu.tw) programs. The possible presence of transmembrane domains in the ORFs analyzed was predicted by TMHMM (www.cbs.dtu.dk/services/TMHMM).
3 Results and discussion
3.1 Electrophoresis of extracellular proteins of A. ferrooxidans grown on sulfur and ferrous sulfate
The 1-D and 2-D electrophoregrams of the extracellular proteins of A. ferrooxidans grown on sulfur and ferrous sulfate are shown in Figs.1(a) and (b), respectively. It was found that the extracellular proteins of A. ferrooxidans grown on sulfur were much more complex than those of A. ferrooxidans grown on ferrous sulfate. Some extracellular polymeric substances were actively expressed when A. ferrooxidans cells were grown on sulfur, and the sulfur became bio-modified when the sulfur was treated by A. ferrooxidans cells[9]. So it is reasonable to infer that extracellular polymeric substances are responsible for overcoming the hydrophobic barrier during colonization of sulfur, and the extracellular proteins have the possible role to bond the sulfur with the direct biochemical reaction.
3.2 Identification and predication of extracellular proteins spots
As shown in Fig.1, there were some protein spots with relatively high abundance in sulfur energy substrate but not in ferrous sulfate energy substrate. Eighteen protein spots with relatively high abundance in sulfur substrate were excised in gel. Identification and predication of these eighteen extracellular proteins were made by MALDI-TOF/TOF followed by database searches against SwissProt and AFE-3217 based on the peptide mass fingerprints, and the results are listed in Table 1.
Table 1 Identification and predication of extracellular proteins spots of A. ferrooxidans in sulfur energy substrate by MALDI-TOF/ TOF followed by database searches against SwissProt and AFE-3217 based on peptide mass fingerprints
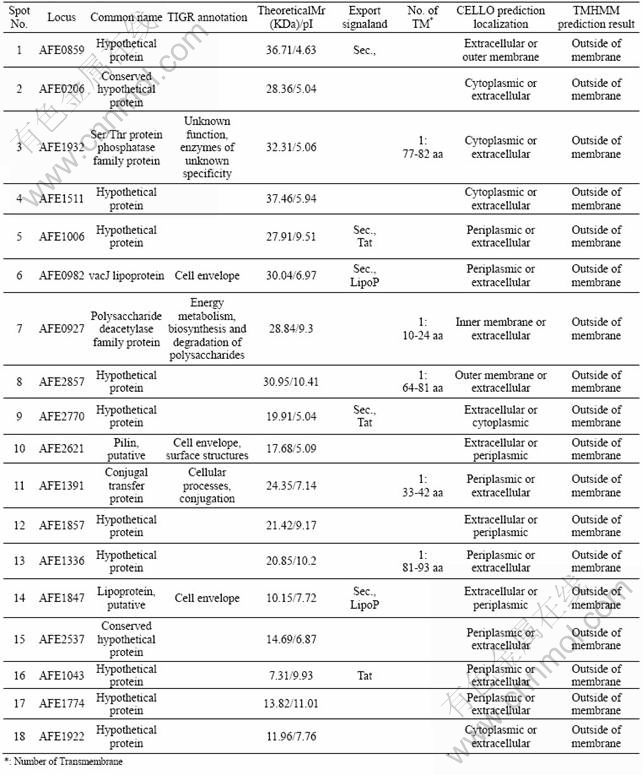

Fig.1 Extracellular proteins of A. ferrooxidans grown on sulfur and ferrous sulfate separated by 1-D (a) and 2-D (b) electrophoresis
3.3 Classification and analysis of extracellular proteins of A. ferrooxidans in elemental sulfur
The identification and predication of extracellular protein spots were divided into different categories according to their potential biological functions in the cell, i.e., AFE_1391: conjugal transfer protein; AFE_2621: pilin; AFE_1847: lipoprotein; AFE_0982: vacJ lipoprotein; AFE_0927: polysaccharide deacetylase family protein; AFE_1932: Ser/Thr protein phosphatase family protein and hypothetical proteins. The percents of each classified extracellular protein category of A. ferrooxidans grown on elemental sulfur are shown in Fig.2.
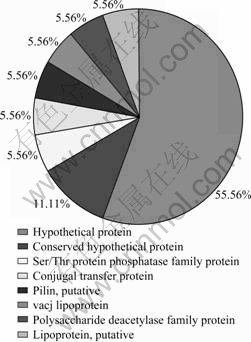
Fig.2 Percents of each classified extracellular protein category of A. ferrooxidans grown on elemental sulfur
All the 18 identified extracellular proteins of A. ferrooxidans grown on elemental sulfur are predicted by their extracellular subcellular localization. Most of the identified proteins are hypothetical proteins (12 of the 18) which are not functionally classified in the TIGR database (www.tigr.com). Only six identified proteins have their own known functions according to the classification in TIGR database. One conjugal transfer protein (conjugal transfer protein: AFE_1391) distributes on the surface of the bacteria, and it performs an important role in gene transformation and recombination through contacting with bacteria, especially the transformation of resistance plasmid by the conjugal transfer protein, which can improve the ability of the bacteria grown in adverse circumstances.
The pilus protein (AFE_2621: pilin, putative, cell envelope, surface structures), which is classified to the cell envelope proteins based on functional prediction, takes part in the composition of cell surface structure. Common pilus gives many kinds of Gram-negative bacteria the ability of adherence and ecesis on various cell surface, which is related to the pathogenicity of some bacteria[15]. The elemental sulfur has a hydrophobic solid surface. The presence of pilus protein in the present experiment may be explained by the bacteria adsorption to the sulfur solid particles, just like the adsorption of pathogenic bacteria to the cell.
There are two kinds of extracellular proteins of the typical signal peptide sequence of lipoproteins (AFE_1847: lipoprotein, putative, cell envelope; AFE_0982: vacJ lipoprotein, cell envelope). vacJ lipoprotein is an extracellularly anchored protein necessary for the Shigella flexneri transmission between bacteria[16]. The function of presence of vacJ lipoprotein during the process of A. ferrooxidans grown on sulfur remains to uncover.
The polysaccharide deacetylase family protein (AFE_0927) and Ser/Thr protein phosphatase family protein (AFE_1932) do not have short signal peptide fragments, but each peptide has a section of transmembrane peptide fragment. They may take part in the synthesis of polysaccharide and signal transduction of cell surface, respectively.
Interestingly, 12 of the 18 proteins are all unknown proteins. Some of these unknown proteins don’t have the characteristics of signal peptide sequence, neither the transmembrane features. Such short peptide that localizes in the extracellular may be secreted to the extracellular space by the non-classical secretory pathway[17]. Additionally, six of these peptide sequences of the low molecular mass (AFE_1336, AFE_1847, AFE_2537, AFE_1043, AFE_1774 and AFE_1922) contain abundant of cysteine residues (Cys), and four of the six proteins have CXXC functional motif.
The thiol groups and the CXXC functional motif are the important active sites of proteins in biochemical reactions. The extracellular hydrophilic peptides abundant of cysteine residues may play a very important role in the chelation with heavy metals, protein modification, stress tolerance, signal transduction and other functions. Many redox proteins are involved in the formation of disulfide bonds. Many oxidation-reduction reactions in organisms are mediated by the thiol oxidoreductase that contains CXXC functional motif as active sites. Protein structure can change also through the conversion between thiol group (—SH) and disulfide bond (—SS—), thus affecting the activity and function of protein molecules.
On the sulfur bio-oxidation, first of all, the acidophilic sulfur-oxidizing bacteria are required to contact and adsorb to sulfur. The effective absorption of bacteria determines an efficient oxidation on sulfur particles. The bacterial attachment to sulfur particles is attained by active expressing of some extracellular polymers substance, and the most important change is that the sulfur particle becomes bio-modified through the biochemical reactions between bacterial extracellular proteins and sulfur. The bio-sulfur is the available energy material for A. ferrooxidans cells. It is no doubt that these biochemical reactions are dominated by the extracellular proteins. It is reported that the sulfur must be used on the premise of its activation, but this step relies on the existence of low-molecular-mass mercaptan (such as glutathione: GSH) [8, 16, 18]. The proteins rich in thiol groups on A. ferrooxidans cell surface may be directly involved in the sulfur activation by using their thiol groups (Pr-SH) to bond sulfur and become Pr-Sn-SH. This assumption, however, needs to prove by further researches.
4 Conclusions
1) The fractions of the extracellular proteins of A. ferrooxidans grown on elemental sulfur and ferrous sulfate were selectively prepared and separated by 2-D electrophoresis. The general pattern of the extracellular proteins for sulfur-grown A. ferrooxidans showed obvious differences from that observed for the ferrous iron-grown bacterium.
2) Some protein spots with apparent higher abundance in sulfur energy substrate were identified by using MALDI-TOF/TOF and the extracellular proteins were classified according to their functions as conjugal transfer protein, pilin, vacJ lipoprotein, polysaccharide deacetylase family protein, Ser/Thr protein phosphatase family protein and hypothetical proteins. Several extracellular proteins abundant of thiol groups and with CXXC functional motif may be directly involved in the sulfur activation by use of their thiol group (Pr-SH) to bond the elemental sulfur and form Pr-Sn-SH.
References
[1] ROHWERDER T, GEHRKE T, KINZLER K, SAND W. Bioleaching review (part A): Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation [J]. Applied Microbiology and Biotechnology, 2003, 63(2/3): 239-248.
[2] SCHIPPERS A, SAND W. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur [J]. Applied and Environmental Microbiology, 1999, 65(1): 319-321.
[3] KNICKERBOCKER C, NORDSTROM D K, SOUTHAM G. The role of "blebbing" in overcoming the hydrophobic barrier during biooxidation of elemental sulfur by Thiobacillus thiooxidans [J]. Chemical Geology, 2000, 169(3/4): 425-433.
[4] CHI A, VALENZUELA L, BEARD S, J MACKEY A, SHABANOWITZ J, HUNT D F, JEREZ C A. Periplasmic proteins of the extremophile Acidithiobacillus ferrooxidans: A high throughput proteomic analysis [J]. Molecular and Cellular Proteomics, 2007, 6(12): 2239-2251.
[5] ROHWERDER T, SAND W. Oxidation of inorganic sulfur compounds in acidophilic prokaryotes [J]. Engineering in Life Sciences, 2007, 7: 301-309.
[6] GEHRKE T, TELEGDI J, THIERRY D, SAND W. Importance of extracellular polymeric substances from Thiobacillus ferrooxidans for bioleaching [J]. Applied and Environmental Microbiology, 1998, 64(7): 2743-2747.
[7] SAND W, GEHRKE T. Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron (III) ions and acidophilic bacteria [J]. Research in Microbiology, 2006, 157: 49-56.
[8] ROHWERDER T, SAND W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and acidiphilium spp [J]. Microbiology, 2003, 149(7): 1699-1710.
[9] ZHANG Cheng-gui, XIA Jin-lan, DING Jian-nan, OUYANG Xu-dong, NIE Zhen-yuan, QIU Guan-zhou. Cellular acclimation of Acidithiobacillus ferrooxidans to sulfur bio-oxidation [J]. Mineral and metullurgical Processing Journal, 2009, 26(1): 30-34. (In press)
[10] OHMURA N, TSUGITA K, KOIZUMI J I, SAIKI H. Sulfur-binding protein of flagella of Thiobacillus ferrooxidans [J]. Applied and Environmental Microbiology, 1996, 178(19): 5776-5780.
[11] BUONFIGLIO V, POLIDORO M, SOYER F, VALENTI P, SHIVELY J. A novel gene encoding a sulfur-regulated outer membrane protein in Thiobacillus ferrooxidans [J]. Journal of Biotechnology, 1999, 72(1/2): 85-93.
[12] BUONFIGLIO V, POLIDORO M, FLORA L, CITRO G, VALENTI P, ORSI N. Identification of two surface proteins involved in the oxidation of sulphur compounds in Thiobacillus ferrooxidans [J]. FEMS Microbiology Reviews, 1993, 11(1/3): 43-50.
[13] SRISOMSAP C, SAWANGAREETRAKUL P, SUBHASITANONT P, PANICHAKUL T, PANICHAKUL T, KEERATICHAMROEN S, LIRDPRAPAMONGKOL K. Proteomic analysis of cholangiocarcinoma cell line [J]. Proteomics, 2000, 4(4): 1135-1144.
[14] HANNA S L, SHERMAN N E, KINTER M T, GOLDBERG J B. Comparison of proteins expressed by pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: An analysis by 2-D gel electrophoresis and capillary column [J]. Microbiology, 2000, 146(10): 2495-2508.
[15] JOHNSON J R, STELL A L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise [J]. Journal of Infectious Diseases, 2000, 181(1): 261-272.
[16] SUZUKI T, MURAI T, FUKUDA I, YOSHIKAWA M, SASAKAWA C. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri [J]. Molecular Microbiology, 1994, 11(1): 31-41.
[17] DRIESSEN A J, FEKKES P, VAN DER WOLK J P. The Sec system [J]. Current Opinion in Microbiology, 1998, 1(2): 216-222.
[18] SUZUKI I. Oxidation of elemental sulfur by an enzyme system of Thiobacillus thiooxidans [J]. Biochimica et Biophysica Acta, 1965, 104(2): 359--371.
Foundation item: Project(50674101) supported by the National Natural Science Foundation of China; Project(2004CB619201) supported by the National Basic Research Program of China; Project(50621063) supported by the Chinese Science Foundation for Distinguished Group
Corresponding author: XIA Jin-lan; Tel: +86-731-8836944; E-mail: jlxia@mail.csu.edu.cn
(Edited by YANG Bing)


