
Isolation, characterization and extraction of mer gene of Hg2+ resisting strain D2
ZENG Xiao-xi(曾晓希)1, 2, TAGN Jian-xin(汤建新)2, JIANG Pei(蒋 佩)2,
LIU Hong-wei(刘宏伟)1, DAI Zhi-min(戴志敏)1, LIU Xue-duan(刘学端)1
1. School of Mineral Processing and Bioengineering, Central South University, Changsha 410083, China;
2. Hunan Key Laboratory of Green Packaging and Biological Nanotechnology,Hunan University of Technology, Zhuzhou 412007, China
Received 4 March 2009; accepted 31 December 2009
Abstract:
Mercury-resistant strain D2 was isolated from mercury-contaminated soil and investigations on its 16S rDNA sequence, growth, minimal inhibitory concentrations (MICs) of metals, antibiotic susceptibility and mer gene were conducted. The strain D2 can grow in the medium containing 60 mg/L mercury ion. It presents more than 99% identity of 16S rRNA gene with Pseudomonas aeruginosa, and exhibits high MIC values for heavy metals and a large spectrum antibiotics resistance. The mer RT gene sequence was amplified from chromosome. Strain D2 is identified as Pseudomonas aeruginosa and the resistance to mercuric ion is related to chromosome.
Key words:
isolation; mercury-resistant strain; Pseudomonas aeruginosa; mer RT;
1 Introduction
As a highly toxic heavy metal, mercury has been widely used with such properties as unique conductivity, ability to form alloys with many metals, uniform volume expansion, and highly efficient catalysis. At present, mercury pollution in the environment mainly comes from mercury mining, gold smelting, fuel combustion, instrument manufacturing, chloride production, antiseptics, fungicides, bactericidal agents and so on[1-2]. Mercury has posed a serious threat to human, animals and plants[3-5].
Due to prolonged exposure to mercury-polluted environment, certain environmental strains of bacteria have acquired highly specific resistance to mercury ion, organ mercury, antibiotics and other heavy metals. They have been paid much attention worldwidely. Mercury-resistant bacteria were isolated from mercury-polluted sites. KAFILZADEH and MIRZAEI[6] found that high mercury levels in the environment can increase the ability of resistance to mercury among the bacterial communities residing in the contaminated sites. Some researchers have focused on the mercury-resistant mechanism. The generally accepted mechanism is that the resistant bacteria have mercuric reductase, and mercury ion can be reduced to Hg0[7-8]. Mercury metal can volatilize out of the system due to its high vapor pressure. Thus, the resistant bacteria can contribute to mercury removal. Moreover, researchers have used mercury-resistant bacteria in bioremediation. Mercury-resistant bacteria detoxification system for mercury was highly effective in chloral kali wastewater and the level of mercury removal can reach 98%[9]. Several marine bacteria highly resistant to mercury can not only detoxify mercury but also remove more than 70% of Cd and 98% of Pb within 72 and 96 h from the growth medium containing 100 mg/L heavy metal[10]. It was confirmed that mercury-resistant bacteria have potential in heavy metals removal. But from the above reports, it is easy to find that the capacity of mercury resistance is still low. In order to enhance the efficiency of Hg2+ removal, it is important to isolate more highly mercury-resistant bacteria. A mercury-resistant bacterium from the polluted soil is isolated and identified. The resistance to heavy metals and antibiotics is examined. The partial mercury-resistant gene is amplified and analyzed for the mercury resistant mechanism.
2 Experimental
2.1 Screening strain
The soil samples were collected from various workshops surrounding Hunan Zhuzhou Smelter, China, where heavy metals have been refined, alloys have been produced for more than 50 years and the soil around it has been seriously polluted. The average mercury ion concentration of the soils is 3.29 mg/kg. 1 000 mg/L stock solution of mercury ion was prepared by dissolving HgCl2 in deionized water and sterilized by using 0.22 μm pore-size sterile filters.
Samples were added to liquid medium with 10 mg/L Hg2+, cultivated at 30 ℃ and 180 r/min in a rotary shaker for 3-4 d. The cultures were plated onto the solid media with Hg2+ from 10 mg/L to 65 mg/L to select the highest mercury-resistant isolate. The microorganism resistance to the highest concentration mercury ion was selected and used in subsequent experiments. In order to obtain purified bacterium, single colony was picked and streaked on the solid medium. This process was repeated more than three times.
2.2 Analysis of 16S rDNA
The genomic DNA was extracted using Bacteria Genomic DNA Extraction Kit (TIANamp Bacteria, TIANGEN) and the amplification reaction was performed using the universal 16S rDNA primers 27f (5’-GAGAGTTTGATCCTGGCTCAG) and 1521r (5’-AAGGAGGTGATCCAGCC). PCR reaction was performed as follows: 95 ℃ for 3 min; 32 cycles of 94 ℃ for 1 min, 56 ℃ for 1 min, and 72 ℃ for 2 min; and 72 ℃ for 10 min. The PCR product was purified with a Gel Extraction Kit and sequenced in Shanghai Sangon Biotechnologies Co. Ltd., China.
16S rDNA sequence was submitted to GenBank and compared with similar sequences by BLAST analysis. Phylogenetic tree derived from 16S rDNA was constructed using the soft ware Clustal W.
2.3 Effect of mercuric ion on bacterial growth
The growth kinetic was determined in Beef extract-peptone medium in the present and absence of Hg2+. The isolate was inoculated in flasks with mercury ion concentrations of 10, 30 and 50 mg/L. The flasks were incubated on a rotary shaker at 30℃ and 180 r/min. The cultures from all the flasks were measured at a regular interval of 660 nm using a spectrophotometer. The absorption values were plotted to draw growth curves. The bacterium cultured in the absence of mercuric ion was used as a control.
2.4 Resistance to other heavy metals
The resistance to other heavy metals was tested for MIC. Stock solutions of other heavy metals including Cd(NO3)2, CuCl2, CoCl2, ZnSO4, MnSO4 and Pb(Ac)2 were prepared in the same manner of mercury ion solution. The strain was streaked on Beef extract-peptone agar plates with single metal. The control experiment was carried out on E .coli conserved in the laboratory.
2.5 Resistance to antibiotics
The 12 antibiotic disks (purchased from Hangzhou Microbe Reagent Limited Company, China) were used in this test: penicillin (6 μg), ampicillin (10 μg), amikacin (30 μg), streptomycin (10 μg), neomycin (30 μg), tetracycline (30 μg), erythromycin (15 μg), kitasamycin 15 μg), nalidixic acid (30 μg), polymyxin (36 μg), novobiocin (30 μg), and chloramphenicol (30 μg). The strain was determined to be sensitive or resistant to the 12 antibiotics. 0.1 mL culture was plated onto beef extract-peptone agar plates. 2-3 antibiotic disks were placed on each plate and incubated at 30 ℃ for 3-5 d.
2.6 Amplification of mercury resistance gene
Mercury-resistant gene was amplified by the following primers: mer 1, 5’-GAGATCTAAAGCACG- CTAAGGC; mer 2, 5’-GGAATCTTGACTGTGATCG- GG[11]. PCR reaction was performed as follows: 95 ℃ for 5 min; 32 cycles of 94 ℃ for 30 s, 52 ℃ for 30 s, 72 ℃ for 1 min; and 72 ℃ for 7 min. PCR product was purified with a Gel Extraction Kit and sent to Shanghai Sangon Biotechnologies Co. Ltd., China, to sequence. Mer gene sequence was submitted to GenBank and compared with similar sequences by BLAST analysis.
3 Results and discussion
3.1 Isolation and identification of mercury-resistant bacteria
A strain, named as D2, which could tolerate 60 mg/L mercuric ion, was isolated from soil samples. It was Gram-negative and rod-shaped. 16S rDNA sequence of strain D2 was submitted to the database of GenBank and the submission number is EU915713. The BLAST analysis shows the partial 16S rDNA of D2 had more than 99% identity with that of P. aeruginosa. The phylogenetic tree based on 16S rDNA was constructed to determine the relationship between strain D2 and other P. aeruginosa (Fig.1). Based the above characterization, strain D2 was identified as P. aeruginosa.
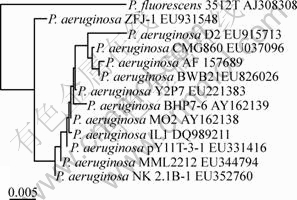
Fig.1 Phylogenetic tree derived from 16S rDNA sequence data of strain D2
Microorganisms might develop various mechanisms to resist antibiotics and tolerate metals under environmental conditions of heavy metal. The presence of such mercury-resistant microorganism is often correlated with the level of mercury contamination in an environment. Strain D2 was isolated from Hunan Zhuzhou Smelter, China, which was polluted with high concentration of mercuric ion. It exhibited high resistance (60 mg/L) to mercuric ion and somewhat higher than reported mercury-resistant isolates[12-13]. Moreover, P. aeruginosa, widely existing in environment, is easy to be cultured and the growth conditions are ordinary. So, with the ability of high resistance, strain D2 has the potential of application in recovering mercuric pollution environment.
3.2 Effect of mercuric ion on bacterial growth
Heavy metals may exert an inhibitory action on microorganisms by blocking essential functional groups, displacing essential metal ions, or changing the active conformations of biological molecules. The mechanism of resistance to metal includes two types: accumulation in the form of particular protein-metal association and blockage at the level of the cell wall and the systems of membrane transportation[14]. Generally, the toxicity to microorganism increases with the increase of heavy metals concentration. The growth responses of strain D2 to different concentrations of mercury ion in liquid cultures are given in Fig.2. Compared with the control group without mercury ion medium, mercury ion had obvious toxicity to cells. With the increase of mercury ion concentration in medium, the strain exhibited a longer lag phase and lower final cell density. Strain D2 can grow in solid medium with 60 mg/L mercuric ion, but in liquid medium containing 60 mg/L mercuric ion, it grew slowly and the final cell density was so low that the culture of shake flasks almost kept transparent. It is clear that when the concentration of mercuric ion in liquid medium reaches 60 mg/L, the toxicity to cell is very serious and the bacterium may not proliferate. Investigation of growth characteristics with mercury ion medium may be of practical interest to application in bioremediation.
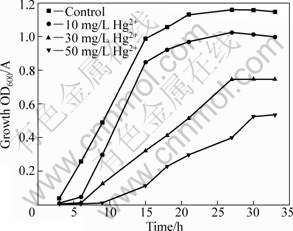
Fig.2 Growth curves of strain D2 in media containing different concentrations of mercury ion
3.3 Resistance to other heavy metals
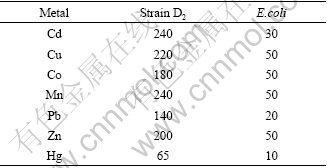
The MIC (minimal inhibitory concentration) means the lowest concentration of metal that completely prevented bacterium growth. Cu, Co, Mn and Zn are essential for bacterium as trace nutrients; and Cd, Hg and Pb have no known beneficial roles. MICs of strain D2 to seven heavy metals are presented in Table 1. In comparison with other six heavy metals, the MIC of mercury was the lowest in this study, but it was higher than the bacteria in other reports[12-13]. It can be concluded that strain D2 can resist to all the metals, which may be related with the sample site. Resistance to various heavy metals is an advantage for bacterium while performing the desired mercury reduction in the heavy metals polluted environment.
Table 1 MIC of strain D2 to various heavy metals (mg/L)
3.4 Antibiotic susceptibility
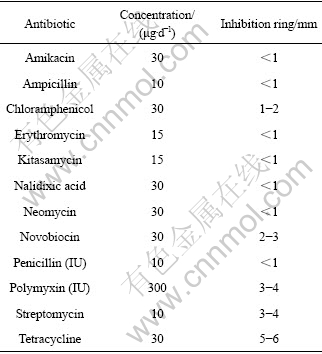
After incubating for 36-48 h, colonies appeared on the plates with antibiotics disk. On the plates inoculated with strain D2, except that the plates with polymyxin, novobiocin, streptomycin and tetracycline exhibited 2-6 mm inhibition ring, there was no obvious inhibition ring on the other plates. According to the Ref.[15], strain D2 has resistance to the 12 antibiotics, as listed in Table 2.
Table 2 Inhibition ring of strain D2 to antibiotics
3.5 Amplification and analysis mer gene
The mer gene sequence was obtained by polymerase china reaction using the primer pair mer 1 and mer 2 (Fig.3). The sequence was submitted to GenBank (Accession FJ230967). Blast in Genbank, the sequence has more than 99% similarity to the sequences from P. aeruginosa, P. putida, P.fluorescens, R. metallidurans and so on. Among the similar sequences, Tn 501 and the plasmids pMOL28, pMOL30 were well studied about heavy metals resistance[16]. Compared with those by using the multiple sequences alignment program Clustal W, the query sequence from the isolated strain D2 exhibited almost 100% identity with sequence in the database (Fig.4). From the analysis of the genes sequences, the sequence obtained from strain D2 chromosome was estimated to be encoded with the genes mer T (112-462 bp) and mer R (534-966 bp).
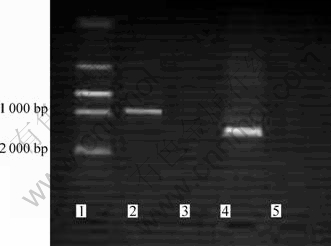
Fig.3 Agarose gel electrophoresis of PCR produces (1—DNA size marker; 2—Mer gene; 4—16S rDNA; 3, 5—Control reaction without adding template DNA)
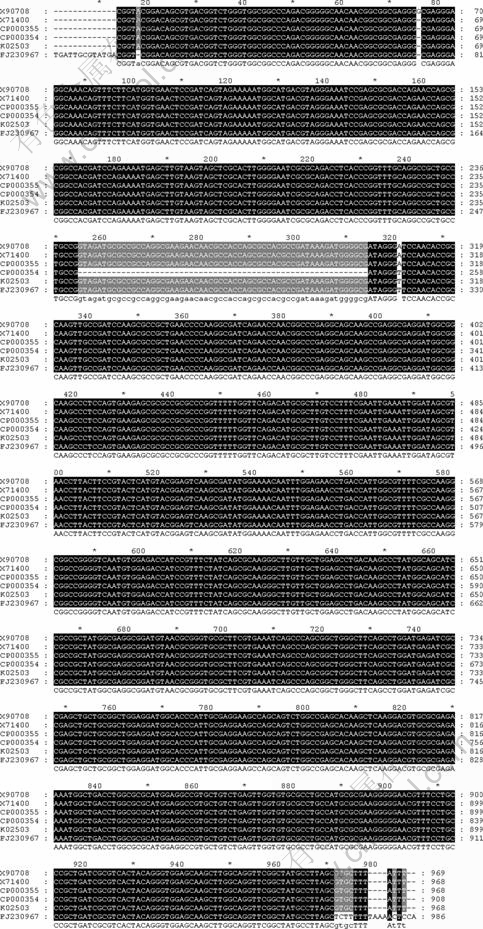
Fig.4 Sequences alignment (FJ230967: mer gene strain D2 sequence; K02503: P. aeruginosa transposon Tn501 mercuric ion resistance operon; X71400: R.metallidurans CH34 plasmid pMOL30; CP000354: R. metallidurans CH34 plasmid pMOL30, complete sequence; X90708: R. metallidurans CH34 plasmid pMOL28; CP000355: R. metallidurans CH34 plasmid pMOL28, complete sequence)
It is well known that the reduction is conferred by cluster of the genes organized in an operon called mer operon. The mer operon can be divided into narrow-spectrum mercury-resistant operon and broad-spectrum mercury-resistant operon. Narrow- spectrum mercury operon confers resistance to Hg2+ and typically contains mer R and structural gene (mer TPCAD). Mer R is transcribed divergently from the other mer genes and encodes a repressor/activator protein. Mer TP gene encodes a transport system (in some cases, an additional mer C gene). Mer A encodes the mercuric ion reductase, and mer D involves in down-regulation of the operon. Strain D2 can be resistant to inorganic mercury but not to organomercurials, so it belongs to narrow-spectrum mercury-resistant one.
The mer operons contain the essential genes: mer A, mer R, mer T, mer P and mer B. These mer operons are often localized on plasmids and other mobile elements, such as transposons. Transposons carrying mer operons have been identified from both clinical and environmental bacteria. Tn501 is the widely investigated mercury-resistant transposon[17]. In this study, the mer RT genes were amplified from chromosome and they are highly similar to plasmid and transposon. It is demonstrated that mer RT are highly conserved in mercury-resistant bacteria and some of mercury- resistant determinants have moved from plasmid to chromosome. Strain D2 mercury resistance is chromosomally located. The result agrees with the studies in Ref.[18].
4 Conclusions1) A strain D2 resistant to high concentration mercury ion was isolated from heavy metals polluted soil. Based on analysis of 16S rDNA, the strain was identified as P. aeruginosa. It can grow in the soil medium containing 60 mg/L Hg2+. But when the concentration of mercuric ion in liquid medium reaches 60 mg/L, the strain growth almost stops.
2) Besides mercury, strain D2 can tolerant to other heavy metals Cu, Co, Mn, Zn Cd, and Pb. Antibiotics disc tests demonstrate it could be resistant to the 12 antibiotics: penicillin, ampicillin, amikacin, streptomycin, neomycin, tetracycline, erythromycin, kitasamycin, nalidixic acid, polymyxin, novobiocin and chloramphenicol.
3) The mer gene RT sequence was obtained by primer pair mer 1 and mer 2. Sequence alignment demonstrates that the obtained mer gene might encode mer RT. The resistance to mercuric ion of strain D2 was related to chromosome.
References
[1] MORENO F N, ANDERSON C W N, STEWART R B, ROBINSON B H. Phytofiltration of mercury-contaminated water: Volatilisation and plant-accumulation aspects [J]. Environmental and Experimental Botany, 2008, 62: 78-85.
[2] KANNAN S K, KRISHNAMOORTHY R. Isolation of mercury resistant bacteria and influence of abiotic factors on bioavailability of mercury—A case study in Pulicat Lake North of Chennai, South East India [J]. Science of the Total Environment, 2006, 367: 341-353.
[3] BRIDGES C C, ZALUPS R K. Homocysteine, system b0, and the renal epithelial transport and toxicity of inorganic mercury [J]. American Journal of Pathology, 2004, 165(4): 1385-1394.
[4] RUDOLFS K, ZALUP S, SARFARAZ A. Homocysteine and the renal epithelial transport and toxicity of inorganic mercury: Role of basolateral transporter organic anion transporter 1 [J]. J Am Soc Nephrol, 2004, 15: 2023-2031.
[5] PATRA M, BHOWMIK N, BANDOPADHYAY B, SHARMA A. Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance [J]. Environmental and Experimental Botany, 2004, 52: 199-223.
[6] KAFILZADEH F, MIRZAEI N. Growth pattern of Hg resistant bacteria isolated from Kor River in the presence of mercuric chloride [J]. Pak J Biol Sci, 2008, 11(18): 2243-2248.
[7] HOLTZE M S, NIELSEN P, EKELUND F, RASMUSSEN L D, JOHNSEN K. Mercury affects the distribution of culturable species of pseudomonas in soil [J]. Applied Soil Ecology, 2006, 31: 228-238.
[8] TAKEUCHI F, NEGISHI A, MAEDA T, KAMIMURA K, SUGIO T. Volatilization and recovery of mercury from mercury wastewater produced in the course of laboratory work using Acidithiobacillus ferrooxidans sug 2-2 cells [J]. Journal of Bioscience and Bioengineering, 2003,95(3): 239-244.
[9] von CANSTEIN H, LI Y, TIMMIS K N, DECKWER W D, WAGNER-D?BLER I. Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant pseudomonas putida strain [J]. Applied and Environmental Microbiology, 1999, 65(12): 5279-5284.
[10] DE J, RAMAIAH N, VARDANYAN L. Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury [J]. Mar Biotechnol, 2008, 10(4): 471-477.
[11] ABOU-SHANAB R A I, BERKUM P, ANGLE J S. Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale [J]. Chemosphere, 2007, 68(2): 360-367.
[12] DE J, RAMAIAH N. Characterization of marine bacteria highly resistant to mercury exhibiting multiple resistances to toxic chemicals [J]. Ecological Indicators, 2007, 7: 511-520.
[13] GUPTA A, RAI V, BAGDWAL N, GOEL R. In situ characterization of mercury-resistant growth-promoting fluorescent pseudomonads [J]. Microbiological Research, 2005, 160: 385-388.
[14] HASSEN A, SAIDI N, CHERIF M, BOUDABOUS A. Resistance of environmental bacteria to heavy metals [J]. Bioresource Technology, 1998, 64: 7-15.
[15] DONG X, CAI M. Manual of common determinative bacteriology [M]. Beijing: Science Press, 2001. (in Chinese)
[16] READY D, PRATTEN J, MORDAN N, WATTS E, WILSON M. The effect of amalgam exposure on mercury- and antibiotic-resistant bacteria [J]. International Journal of Antimicrobial Agents, 2007, 30: 34-39.
[17] MONCHY S, BENOTMANE M A, JANSSEN P, VALLAEYS T, TAGHAVI S, LELIE D, MERGEAY M. Plasmids pMOL28 and pMOL30 of cupriavidus metallidurans are specialized in the maximal viable response to heavy metals [J]. J Bacterial, 2007, 189: 7417-7425.
[18] BARBIERI P, BESTETTI G, RENIERO D, GALLI E. Mercury resistance in aromatic compound degrading pseudomonas strains [J]. FEMS Microbiology Ecology, 1996, 20: 185-194.
Foundation item: Project(50621063) supported by the National Natural Science Foundation of China; Project (2004CB619204) supported by the National Basic Research Program of China; Project(2009sk3035) supported by the Science and Technology Program of Hunan Province, China
Corresponding author: LIU Xue-duan; Tel: +86-731-88836019; E-mail: xueduanliu@sina.com
DOI: 10.1016/S1003-6326(09)60170-9


