- Abstract:
- 1 Introduction▲
- 2 Experimental▲
- 3 Results and discussion▲
- 4 Conclusions▲
- References
- Figure
- Fig.1 Morphological survey results of strain DS-0205: (a) Colony photograph on agar culture; (b) DS-0205 morphology under optical microscope; (c) Results of capsule stain; (d) Ultra-microstructure under SEM; (e) Ultra-microstructure under TEM
- Fig.2 Phylogenetic positions of deep-sea strain DS-0205 within Sporidiobolus clade (tree is constructed by the maximum likelihood method described in text): (a) On the basis of a total of 1 428 aligned nucleotide sites in 18S rRNA; (b) On the basis of a total of 588 aligned nucleotide sites in ITS-5.8S rRNA sequences
J. Cent. South Univ. Technol. (2009) 16: 0942-0947
DOI: 10.1007/s11771-009-0157-5
![]()
Isolation and identification of Rhodosporidium diobovatum DS-0205 from
deep-sea sediment of eastern Pacific Ocean
ZENG Le-ping(曾乐平)1, 2, HUANG Ju-fang(黄菊芳)1, QIU Guan-zhou(邱冠周)3, CHU Feng-you(初凤友)2,
CHEN Dan(陈 旦)1, TONG Jian-bin(童建斌)1, LUO Xue-gang(罗学港)1
(1. Xiangya School of Medicine, Central South University, Changsha 410013, China;
2. Key Laboratory of Submarine Geosciences of State Oceanic Administration, Hangzhou 310012, China;
3. School of Resources Processing and Bioengineering, Central South University, Changsha 410083, China)
Abstract:
A facultative heterotrophic strain (DS-0205) was isolated from a deep-sea sediment sample collected at a depth of 5.2 km in the eastern Pacific Ocean, China. Strain DS-0205 is motile helmet-like single cell or pairs, forming hemisphere with the center sunken of variable size. It has a widespread carbon source and nitrogen source, including agarose, citric acid, salicin, D-glucitol nitrate, sodium nitrite and ethylamine. It can grow in the following environment: temperature 4-37 ℃, pH 2.0-12.0, tolerance to NaCl≤15%. Two phylogenetic trees, one based on the ITS and 5.8S rRNA sequences and the other based on the 18S rRNA sequences, unite strain DS-0205(=JCM 0205) to the type strain of Rhodosporidium diobovatum JCM 3787 through a considerable evolutionary distance. These results suggest that strain DS-0205 is a new strain of the Rhodosporidium diobovatum.
Key words:
deep-sea sediment; ITS and 5.8S rRNA sequences; 18S rRNA; isolation; identification;
1 Introduction
During the past thirty years, most scientists have focused on the microorganisms, specifically the bacteria in the sub-seafloor hydrothermal vents, while only a few scientists have studied the microorganisms in the cold deep-sea biosphere[1]. Up to now, the main approach to microorganisms is to study microbial diversity in very niche-specific, whether at vents or elsewhere in the deep ocean[2]. The results have led to new significant discoveries of unusual microbial diversity (cultured and uncultured), metabolic activity, molecular phylogeny, and natural products[3]. Compared with deep-sea bacteria, the yeast in deep-sea biosphere, which occupy a wide variety of niches for their highly versatile physiological adaptations, have largely remained neglected[4].
Recent researches have indicated that a lot of red yeasts which produce carotenoids exist in many benthic sediments and animals in the Pacific Ocean[5]. As an important member among the red yeasts, Rhodosporidium diobovatum, which produces carotenoids, was detected from the marine environment many years ago, but it was rarely isolated from deep-sea environment, especially in 5.0 km benthic environment[6-7]. As is known, it was very difficult to obtain pure strains from deep-sea mud or benthic organisms[8]. Most researches were limited to microorganisms in relatively shallow regions of ocean. However, the environment in the deep ocean was characterized with low temperature, high hydrostatic pressure, high salt and a ‘feast and famine’ nutrient condition. This kind of extreme environment endowed many special properties for deep-sea yeasts, which might possess greater value in economic exploitation than those on the land. Now, increasing interest in the deep sub-seafloor biosphere yeast becomes the foci of the studies on their gene resources, enzymes and protein resources, extracellular protease resources, and so on[9-10]. But pure cultures still remain essential to be recognized, comprehended and exploited.
The aim of this work was to develop methodologies to cultivate environmentally relevant heterotrophic yeast from the deep sub-seafloor biosphere by using different organic carbon concentrations and inoculum dilutions in enrichment cultures. Moreover, to avoid bias during isolation[11], several identification methods were adopted, including morphological, physiological and biochemical methods with 18S rRNA gene and ITS-5.8S rRNA sequence-based techniques[12-14].
2 Experimental
2.1 Sample collection
Deep-sea sediment samples were collected from the eastern Pacific Ocean in China (8.334?N; 145.384?E), at a depth of 5.2 km situ by the China ship Da-Yang 105 in December 2003. The temperature in situ was 4℃, and the pH was 7.60.
2.2 Enrichments and isolation
The culture media[8] were prepared with artificial seawater supplemented with general culturing media, malt agar (5% Fleischmann’s dry diastatic diamalt 20, 0.1% yeast extract, with/without 2% agar), and 0.01% chloramphenicol and 0.002% streptomycin[15].
Firstly, the samples were incubated at a low temperature 4-15 ℃ for 7-10 d. Then the enriched liquid was filtered through sterile 0.22 μm Millipore membrane filters and placed on agar plates for incubation, and a few colonies were produced. Concentration of liquid cultures and subsequent spreading on agar plates were necessary to obtain the isolates.
2.3 Morphological survey
The morphology of strain DS-0205 was characterized using standard methods with tiny modifications[15]. The shape and color of the colonies were observed under macroscopic, and the morphologic characteristics were observed under optical microscope. Scan electron microscope (SEM) and transmission electron microscope (TEM) were used to get the ultra-microstructure of the strain DS-0205.
2.4 Physiological and biochemical characteristics
Strains were characterized physiologically and biochemically by standard methods with some modifications[15]. Assimilation of nitrogen compounds was examined on solid media using starved inocula. The vitamin requirements and its Ubiquinone were investigated according to the Kurtzman’s methods. The temperature range for cell growth was determined using samples (0.2 mL) taken from 10 mL of cultures cultivated overnight. Tubes were incubated at 4, 10, 15, 25, 30, 37 and 42 ℃, respectively. The pH range from 2.0 to 12.0 was tested for cell growth. NaCl tolerance was tested at NaCl concentrations of 0%, 1%, 2%, 5%, 10%, 15% and 20%, respectively, in liquid medium in replicate tubes. Anaerobic test was performed according to the Fell’s method[9]. Growth was recorded daily for 30 d.
2.5 DNA extraction and detecting methods
It was very difficult to extract DNA from the strain DS-0205 because of its thick capsule. In this work, the DNA extraction method presented was a slight modification of that used for bioleaching bacteria[16]. Purified cell pellets about 0.1 g in sterile 5 mL polypropylene centrifuge tube were thoroughly suspended and washed three times in 5 mL autoclaved, high-salinity, phosphate-buffered saline (PBS) (300 mmol NaCl, 2.7 mmol KCl, 10 mmol Na2HPO4, 1.7 mmol NaH2PO4), with a centrifuge at 4 ℃ and 10 000 r/min for 10 min between two washing steps. The cell pellets were suspended in 2 mL extraction buffer, and incubated in boiling water for 15 min with gentle inversions every 3 min. Then, the following steps were the same as those of the DNA extraction method of the bioleaching bacteria[16].
Cell lysis ratio was estimated for the test sample by direct microscopic counts[16] before and after the DNA extraction treatments. All counts were obtained by one investigator. DNA purity was assessed spectro- photometrically by the ratios of A260/A280 and A260/A230[16]. The quality of the extraction was checked on 1.0% (mass fraction) agarose gel, containing 0.8 mg/L ethidium bromide (BET).
2.6 PCR-amplification
The 18S rRNA gene was amplified with the oligonucleotide primers (5′-ACCTGGTTGATCCTGC- CAG-3′, 5′-TGATCCTTCYGCAGGTTCAC-3′)[12]; and their PCR programs were done carefully. The DNA fragments of ITS region (including 5.8 S rRNA) were amplified with a pair of primers[17] ITS1 5′- GTCGTAACAAGGTTTCCGTAGGTG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), and their PCR programs were carried out carefully. The PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining, and then purified by the QIAquick PCR purification kit (Qiagen, Chatsworth, Calif.) according to the manufacturer’s instructions.
2.7 Cloning, sequencing and phylogenetic analysis
The purified 18 S rRNA PCR products were cloned into the pGEM-T Vector (Promega, America) and transformed to TOP10 competent cells (invitrogen, America) for blue-white screening. The positive clones were sequenced after colony PCR check. All sequences were aligned using ClustalX2.0[18].
3 Results and discussion
3.1 Morphological characteristics of strain DS-0205
On agar plates, after 3 or 5 d at 15 ℃, the colonies of strain DS-0205 obtained by deliquation and/or streak culture are light pink, glistening, soft with smooth and entire margin in aerobic condition(Fig.1(a)). However, the colony color turns from white to pink in anaerobic condition. The time for the colony growth is variable. Clear colonies usually appear after 3 or 5 d or even longer. The colony size is highly variable, and sometimes the growth is confluent with each other.
Under optical and/or electron microscope (Figs. 1(b)-(e)), strain DS-0205 is motile helmet-shaped single cell or pairs, forming hemisphere with the center sunken of variable size (the diameter of organism ranges from 1.5 to 5.0 μm and the center sunken from 0.5 to 2.0 μm) (Figs.1(b) and (d)). It has a thick capsule out of the cell wall (Fig.1(c)), especially in stationary phase of growth. In liquid cultures, strain DS-0205 might form visible spores (Fig.1(c)). Observing the internal structure of strain DS-0205, we find that it has a wall with a thickness from 0.3 to 1.0 μm and a ring-like wall in the middle cell, and has a few sap cavities through TEM(Fig.1(e)).
3.2 Physiological and biochemical characteristics of strain DS-0205
The typical physiological and biochemical characteristics of strain DS-0205 are listed in Tables 1 and 2. Growth in lysine or cadaverine is weak and latent. Growth occurs in the presence of 0.01% cycloheximide. Strain DS-0205 doesnot produce starch-like substances, and its major ubiquinone is Q-10. Both the diazonium blue B reaction and urease activity are positive.
Based on the fairly high similarity of morphological, physiological and biochemical properties, strain DS-0205 is identified as Rhodosporidium diobovatum. Although some teliospores may be spheroidal or obovate, no mating occurs and no teliospores are generally cleft, diobovated or diclavated[8]. Strain DS-0205 also produces carotenoids, which are of great significance for human health for their pro-vitamin A activity and nutraceutical potential[20]. However, to some degree, strain DS-0205 (Fig.1(b)) is different from Rhodosporidium diobovatum[8]. For example, the shape of DS-0205 is somewhat different from each other, and it can use galactose or maltose as the sole carbon source (Table 1). In addition, we find that strain DS-0205 has a widespread carbon source, such as agarose, citric acid, salicin and D-glucitol. It can also use various nitrogen sources, including nitrate, sodium nitrite, and ethylamine (Table 2). Stain DS-0205 possesses good environment- accustoming ability, and is tolerant to low temperature (4-37 ℃), wide pH range (pH from 2.0 to 12.0) and high salinity (≤15% NaCl). These special properties may be related with the specific deep-sea environment, such as low temperature, high salt and famine nutrient condition.
3.3 DNA extraction and PCR amplification
Agarose gel electrophoresis of the DNA extracts reveals that most of the DNA extraction fragments are larger than 23 kb. The A260/A230 ratio of DNA extract is 1.63, and the A260/A280 ratio is 1.71. Post-extraction counts are significantly lower than pre-extraction counts, indicating high lysis efficiencies (90.5%). The DNA templates are extracted from strain DS-0205 by the modified DNA extraction method generating good PCR bands for the nuclear 18S rRNA and ITS and 5.8S rRNA sequences, when the crude DNA yields are used.
The 18S rRNA and the ITS-5.8S rRNA sequences
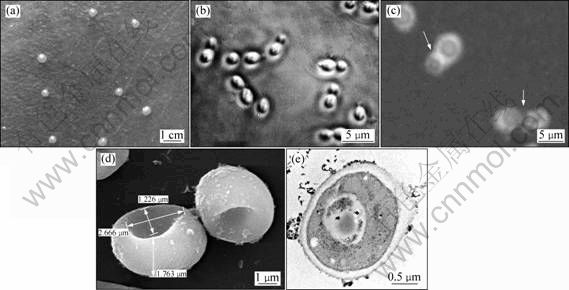
Fig.1 Morphological survey results of strain DS-0205: (a) Colony photograph on agar culture; (b) DS-0205 morphology under optical microscope; (c) Results of capsule stain; (d) Ultra-microstructure under SEM; (e) Ultra-microstructure under TEM
Table 1 Comparison of main characters between strain DS-0205 and Rhodosporidium diobovatum
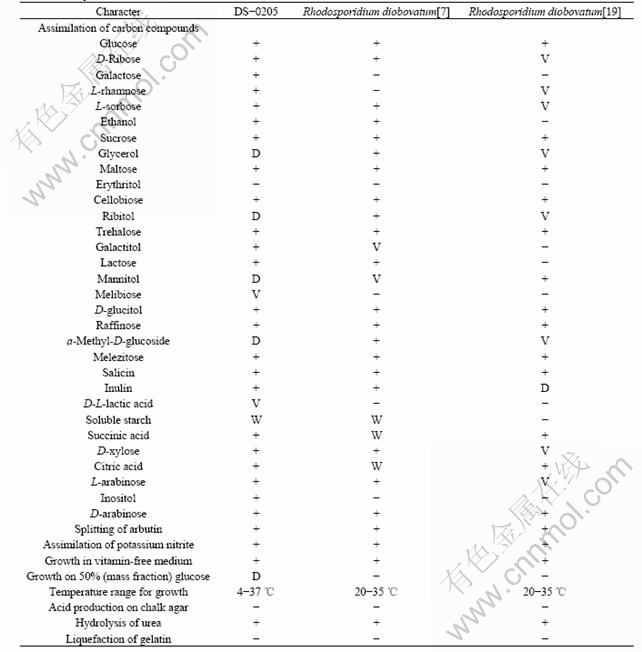
Note: Positive (+), negative (-), weak (W), delayed over 7 d (D), variable (V).
Table 2 Results of utilization of other energy sources for strain DS-0205
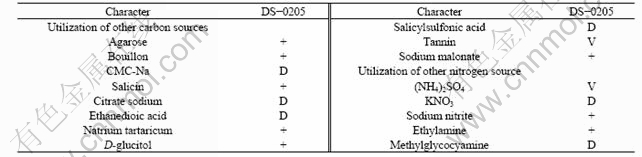
Note: Positive (+), negative (-), weak (W), delayed over 7 d (D), variable (V).
of strain DS-0205 are submitted to GenBank and their assigned accession numbers are DQ854820 (18S rRNA; JCM 0205) and DQ981849 (ITS1-5.8S-ITS2 regions sequence), respectively.
3.4 Phylogenetic analysis
Both ITS-5.8S rRNA gene sequence phylogenetic tree and the 18S rRNA gene sequence phylogenetic tree have united strain DS-0205(=JCM 0205) to strain JCM-3787 of Rhodosporidium diobovatum through a considerable evolutionary distance (Figs.2(a) and (b)).
Sporidiobolus clade represents the red-pigmented teliosporic yeasts: Rhodosporidium and Sporidiobolus with phragmometabasidia, and their relative anamorphs in the genera Rhodotorula and Sporobolomyces. Sporidiobolus clade is distinct from other urediniomycetous yeasts based on the phylogenetic analysis for the 18S rRNA sequences or the sequences of the D1/D2 region of 26S rRNA[21]; nucleotide sequences of internal transcribed spacer (ITS) regions are determined to give guideline for species identification in the genus Rhodosporidium[14]. Thus, to establish the appropriate taxonomic position of strain DS-0205, we determined the sequences of the 18S rRNA, the ITS regions and the 5.8S rRNA. We generated phylogenetic trees of species within the Sporidiobolus clade, adding strain DS-0205, on the basis of either 588 sites in the ITS regions and 5.8S rRNA, or 1 492 sites in the 18S rRNA. Rhodosporidium babjevae and Rhodotorula graminis, were omitted from our analysis, for the 18S rRNA and ITS-5.8S rRNA of these species are very variable compared with those of the other species. Rhodotorula marina JCM 3 776 in Erythrobasidium clade is used as one outgroup.
The 18S rRNA tree is generated by the maximum likelihood (ML) analysis divided the 11 species into three clusters (Fig.2(a)). In the ITS-5.8S rRNA tree (Fig.2(b)), the position of 11 species in respective clusters only changes slightly from the 18S rRNA tree (Fig.2(a)), and positions of cluster 1 and cluster 2 in Fig.2(a) are exchanged as shown in Fig.2(b). The reasons for this position change may be related with base

Fig.2 Phylogenetic positions of deep-sea strain DS-0205 within Sporidiobolus clade (tree is constructed by the maximum likelihood method described in text): (a) On the basis of a total of 1 428 aligned nucleotide sites in 18S rRNA; (b) On the basis of a total of 588 aligned nucleotide sites in ITS-5.8S rRNA sequences
deletion in these strains, all of which possess a 20-41 bases deletion at a site in the ITS1/ITS2 region. The number of deleted bases, 20 bases in the case of DS-0205 and JCM 3787 strains, 28 bases in the case of strain JCM 1814, 32 bases in the case of JCM 8547, CBS 20 and JCM 8171 strains, 34 bases in the case of strain JCM 5296, 39 bases in the case of JCM 10311 and JCM 5296 strains and 41 bases in the case of strain JCM 5350, is unlikely to make no contribution to the phylogenetic relationship inferred from the ITS-5.8S rRNA tree. However, this relationship is essentially supported from phylogenic stakes based on the 18S rRNA. In addition, the topology of the phylogenetic tree based on the 18S rRNA sequences is more similar to that based on the sequences of D1/D2 region of 26S rRNA than to that based on the sequences of the ITS and 5.8S rRNA[22]. The comparison of the two phylogenetic trees within the sporidiobolus clade also shows that strain DS-0205 is a new member of Rhodosporidium diobovatum.
4 Conclusions
(1) A new strain DS-0205 that possesses good environment-accustoming ability, including tolerant temperature (4-37 ℃), acid base (pH from 2.0 to 12.0) and NaCl (≤15%), is isolated from a sediment sample at a depth of 5.2 km in the eastern Pacific Ocean (8.334?N; 145.384?E).
(2) Strain DS-0205 is identified by using morphological characters, physiological and biochemical properties and molecular biologic techniques. All results suggest that strain DS-0205 is a new strain of Rhodosporidium diobovatum.
(3) In this work, a valuable method is developed for cultivating, isolating and identifying deep-sea yeast in laboratory, which lays a solid basis for further recognition, comprehension and exploitation of yeast in deep-sea biosphere.
References
[1] ZHANG Jin-wei, ZENG Run-ying. Analysis and gene cloning of the S-adenosylhomocysteine hydrolase from antarctic psychrotrophilic pseudomonas sp. 7197 strain[J]. Progress in Modern Biomedicine, 2006, 6(4): 5-7. (in Chinese)
[2] HARMSEN H, PRIEUR D, JEANTHON C. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations[J]. Appl Environ Microbiol, 1997, 63(7): 2876-2883.
[3] PACE NORMAN R. A molecular view of microbial diversity and the biosphere[J]. Science, 1997, 276(5313): 734-740.
[4] HUANG Ju-fang, ZENG Le-ping, ZHOU Hong-bo, LIU Xi, LIU Fei-fei, ZHANG Ru-bing. Effect of illumination to the growth of the culturable microorganisms from a deep-sea hydrothermal vent[J]. Biomagnetism, 2005, 5(4): 12-13. (in Chinese)
[5] NAGAHAMA T, HAMAMOTO M, NAKASE T. Rhodotorula benthica sp. nov. and Rhodotorula calyptogenae sp. nov., novel yeast species from animals collected from the deep-sea floor, and Rhodotorula lysiniphila sp. nov., which is related phylogenetically[J]. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(5): 897-903.
[6] GADANHO M, ALMEIDA J M, SAMPAIO J P. Assessment of yeast diversity in a marine environment in the south of portugal by microsatellite-primed PCR[J]. Antonie van Leeuwenhoek, 2003, 84(3): 217-227.
[7] NEWELL S Y, HUNTER I L. Rhodosporidium diobovatum sp. n., the perfect form of an asporogenous yeast (rhodotorula sp.)[J]. Journal of Bacteriology, 1970, 104(1): 503-508.
[8] FELL J W. Yeasts in oceanic regions. Recent advances in aquatic mycology[M]. London: Elek Science, 1976.
[9] van DOVER C L, HUMPHRIS S E, FORNARI D. Biogeography and ecological setting of Indian Ocean hydrothermal vents[J]. Science, 2001, 294(5543): 818-823.
[10] DAMARE S, RAGHUKUMAR C, MURALEEDHARAN U D. Fungi in deep-sea sediments of the central indian basin[J]. Deep-Sea Research I, 2006, 53(1): 14-27.
[11] KARSTEN Z, GERARDO T, MICHAEL R, ELKINS J, MATHURE J, SHORT J M, KELLER M. Cultivating the uncultured[J]. Proc Natl Acad Sci USA, 2002, 99(24): 15681-15686.
[12] SEUNG YEO MOON-VAN D S, RUPERT D W, DANIEL V. Oceanic 18S rRNA sequences from proplankton reveal unsuspected eukaryotic diversity[J]. Letters to Nature, 2001, 409: 606-608.
[13] EMILIE L, CORINNE B, CHRISTOPHE N, CARRIAS J F, VISCOGLIOSI E, AMBLAR D C, TELESPHORE S N. Unveiling fungal zoo flagellates as members of freshwater picoeukaryotes: Evidence from a molecular diversity study in a deep meromictic lake[J]. Environmental Microbiology, 2007, 9(1): 61-71.
[14] HAMAMOTO M, NAGAHAMA T, TAMURA M. Systematic study of basidiomycetous yeasts-evaluation of the ITS regions of rRNA to delimit species of the genus Rhodosporidium[J]. FEMS Yeast Research, 2002, 2(3): 409-413.
[15] KURTZMAN C P, FELL J W. The yeasts: A taxonomic study[M]. 4th ed. New York: Elsevier Amsterdam, 1998.
[16] ZENG Le-ping, HUANG Ju-fang, ZHANG Yan-fei, QIU Guan-zhou, TONG Jian-bin, CHEN Dan, LUO Xue-gang. An effective method of DNA extraction for bioleaching bacteria from acid mine drainage[J]. Applied Microbiology and Biotechnology, 2008, 79(5): 881-888.
[17] CLETUS P K, CHRISTIE J R. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses[J]. FEMS Yeast Research, 2003, 3(4): 417-432.
[18] THOMPSON J D, GIBSON T J, PLEWNIAK F, JEANMOUGIN F, HIGGINS D G. The clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Research, 1997, 25(24): 4876-4882.
[19] BARNETT J A, PAYNE R W, YARROW D. Yeasts: Characteristics and IDENTIFICATION[M]. 3rd ed. Cambridge: Cambridge University Press, 1990.
[20] WRIGHT J A, PIETRANGELO C, MACNAUGHTON A. Influence of simulated upper intestinal parameters on the efficiency of beta carotene micellarisation using an in vitro model of digestion[J]. Food Chemistry, 2008, 107(3): 1253-1260.
[21] FELL J W, BOEKHOUT T, FONSECA A, SCORIETTI G, STATZELL-TALLMAN A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rRNA D1/D2 domain sequence analysis[J]. Int J Syst Evol Microbiol, 2000, 50: 1351-1371.
[22] HAMAMOTO M, NAKASE T. Phylogenetic analysis of the ballistoconidium-forming yeast genus Sporobolomyces based on 18S rRNA sequences[J]. Int J Syst Evol Microbiol, 2000, 50: 1373-1380.
(Edited by YANG You-ping)
Foundation item: Projects(40776035, 40376036) supported by the National Natural Science Foundation of China; Project(DY105-02-04-05) supported by the China Ocean Mineral Resources Research and Development Association; Project(07MX27) supported by the Mittal Student Foundation; Project (2340-74236000003) supported by the Post-graduate Foundation of Hunan Province, China; Project (200805330053) supported by the Doctoral Foundation of Ministry of Education of China
Received date: 2009-01-06; Accepted date: 2009-04-13
Corresponding author: HUANG Ju-fang, Professor; Tel: +86-731-82650426; Fax: +86-731-82650426; E-mail: jufanghuang@yahoo.com.cn
- Isolation and identification of Rhodosporidium diobovatum DS-0205 fromdeep-sea sediment of eastern Pacific Ocean


