
A new strain Leptospirillum ferriphilum YTW315 for bioleaching of metal sulfides ores
ZHANG Rui-yong(张瑞永)1, 2, XIA Jin-lan(夏金兰)1, 2, PENG Juan-hua(彭娟花)1, 2, ZHANG Qian(张 倩)1, 2,
ZHANG Cheng-gui(张成桂)1, 2, NIE Zhen-yuan(聂珍媛)1, 2, QIU Guan-zhou(邱冠周)1, 2
1. School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China;
2. Key Laboratory of Biometallurgy of Ministry of Education, Central South University, Changsha 410083, China
Received 9 December 2008; accepted 25 March 2009
Abstract:
A new strain named YTW315 was isolated from Dexing area using the double-layer culture technique. The morphological, biochemical and physiological characteristics of YTW315 were studied. Physiological investigation indicates that the strain YTW315 is a strict (obligate) chemolithoautotroph, metabolizing ferrous iron and pyrite. The optimal growth conditions for the strain are 40 ℃ and pH 1.6. A phylogenetic analysis based on 16S rRNA sequences shows that the isolate is clustered to Leptospirillum ferriphilum with 99.8% similarity to Leptospirillum ferriphilum strain Fairview and ATCC 49881. The molar fraction of DNA (G+C) of the isolate is 58.1%. The strain can tolerate high concentration of Fe(Ⅲ) and As(V) (500 mmol/L and 50 mmol/L, respectively). Bioleaching experiment indicates that the strain can oxidize Fe(Ⅱ) efficiently, and after 30 d, 44.56% of copper and 95.31% of iron are extracted from chalcopyrite and pyrite, respectively.
Key words:
Leptospirillum ferriphilum; bioleaching; isolation; characterization; phylogenetic analysis;
1 Introduction
Bioleaching is now an economical technology for extraction of metals. In many cases, it offers environmental and technical advantages over other available technologies[1-2]. In the bioleaching systems, the mostly mentioned and researched bacteria are the genus Acidithiobacillus, including mainly A. ferrooxidans, A. thiooxidans, A. albertensis and A. caldus[3-4]. A. ferrooxidans and A. thiooxidans are very often presented in mesophilic bioleaching environment, whereas Leptospirillum species is active in moderately acidothermophilic environment[5]. The important role of leptospirillum in bioleaching of sulfides minerals in acidic environments is well documented[6-7]. Temperatures above 40 ℃ and pH values below 1.0 are more suitable to the growth of Leptospirilla than to the growth of acidithiobacilli[8]. Under these conditions, Leptospirilla have been reported to be important contributors to the generation of acid mine drainage and its associated environmental problems[9]. Leptospirillum species have been recognized as the dominant microbes in the bioreactor processing mineral ores at the temperature above 40 ℃[10]. However, L. ferriphilum, a newly found iron-oxidizing bacterium, has not been well studied[11].
Members of the genus Leptospirillum are small, gram-negative, vibrio- or spiral-shaped cells. They are obligately chemolithotrophic organisms, fixing carbon by the Benson-Calvin cycle, using ferrous iron as their sole electron donor and oxygen as their electron acceptor; and they have been formally recognized as coherent bacteria[12]. The genus Leptospirillum has been divided into three groups, Ⅰ, Ⅱ and Ⅲ, on the basis of 16S rRNA gene phylogeny[13]. Representatives of groups Ⅱ and Ⅲ were identified in the biofilm analyzed by community genomics[14]. L. ferrooxidans is a representative of group Ⅰ and L. ferriphilum is a representative of group Ⅱ. Bacteria of group Ⅱ but none of group Ⅰ were capable of growing at 45 ℃. No cultured representatives of group Ⅲ have been described up to now[15-16].
Cultivating Leptospirillum species in appropriate liquid media is seldom problematic, though there have been numerous reports of difficulties in growing isolates on solid media. Leptospirillum species grow poorly on common solid medium because of the presence of some inhibitor, such as soluble oligosaccharide and monosaccharide produced by the gelling agent agar under acidic conditions[17]. At present, the most successful approach to using laboratory media has been a double-layer plate technique which involves an acidophilic heterotrophic bacterium Acidiphilium SJH. This technique is efficient in isolating the iron-oxidizing bacteria, and it involves incorporating an acidophilic heterotroph in a ferrous iron/Tryptone Soya Broth medium, then covering the set gel with a layer of sterile medium. This solid medium has been proved highly successful and reproducible in growing many strains of L. ferrooxidans. LIU et al[18] used the double layer technique and successfully isolated a vibrio-shaped iron-oxidizing bacterium in China. Also an attempt was made to isolate L. ferriphilum by a double-layer plate technique in which yeast HJM was substituted for Acidiphilium SJH[19].
In this work, by using overlayer technique and adding the Acidiphilium sp. PJH[20] to the underlayer, a new strain Leptospirillum ferriphilum named YTW315 was successfully isolated from acid mine drainages (AMDs) in Jiangxi Province, China. A series of morphological and biochemical characterization as well as the analysis of 16S rRNA sequences were done.
2 Experimental
2.1 Bacteria sample
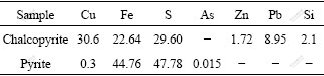
Samples were enrichment culture of acid mine drainages of copper mines in Dexing, Jiangxi Province of China. Chalcopyrite and pyrite used in this experiment were provided by Institute of Mineral Processing Engineering, School of Resources Processing and Bioengineering, Central South University, China. Chemical analysis results of minerals are shown in Table 1.
Table 1 Composition of complex concentrate chalcopyrite and pyrite (mass fraction, %)
2.2 Culture condition
The 9K medium contained (NH4)2SO4 3.0 g/L, KCl 0.1 g/L, K2HPO4 0.5 g/L, MgSO4·7H2O 0.5 g/L, Ca(NO3)2 0.01 g/L.
Solid medium for YTW315 contained two parts: Part Ⅰ, 600 mL 9K liquid medium containing 10 g agar that was autoclaved at 121 ℃ for 20 min; Part Ⅱ, 30 g FeSO4·7H20 dissolved in 400 mL 9K liquid medium, and sterilized by membrane filtration. After equilibrating in water bath at 50 ℃ for 30 min, Part Ⅰ was added to Part Ⅱ and mixed well. The pH was adjusted to 2.0 for plate spreading.
Solid medium for Acidiphilium PJH was follows: 1 L of 9K medium containing 2 g glucose, 0.1 g yeast and 10 g agar. The medium was adjusted at pH 4.5 with concentrated sulfuric acid. This solid medium was used as the underlayer in isolation after cultivation for 1-2 d.
Liquid medium for YTW315 was 9K medium containing 30 g/L FeSO4·7H2O with the initial pH 1.6.
For chemical and genetic tests, cells grown on liquid medium were harvested by centrifugation from a culture at the late-exponential phase of growth, washed twice with sterile 50 mmol/L phosphate buffer (pH 6.8) and pelleted. The cell pellets were used immediately for analysis or stored at -20 ℃ until being analyzed.
2.3 Isolation
The object microorganisms were enriched in the liquid medium, incubated at 40 ℃ and shaken at 180 r/min. The enrichment cultures were transferred to the overlay solid medium and incubated at 35 ℃. After 4-5 d, many colonies formed on the surface of the agar plates. The single colony was picked and streaked on agar plates and incubated at 35 ℃. Agar plate streaks were repeated three times to achieve pure cultures.
2.4 Microscopic studies
Cell motile behavior and Gram staining performed with a Gram stain reagent kit (Haitai Biotech) were observed with Olympus CX 31 optical microscope. Fine morphological features were revealed by transmission scanning microscope (TEM, JEOL JEM-1230) and scanning electron microscope (SEM, JEOL JSM-6360 LV).
2.5 Optimal pH and temperature for growth
Bacteria were suspended in 250 mL flasks containing 100 mL of sterile medium 9K and incubated on a rotary shaker at 250 r/min. To determine the optimum temperature of the culture, it was maintained at pH 1.6 and the temperature was held at set points between 25 ℃ and 55 ℃. To determine the optimum pH for growth, a similar protocol was used except that the culture was incubated at 40 ℃ and with different initial pH values from 0.5 to 3.0.
2.6 Tolerance to heavy metals
The tolerance to some heavy metals of isolate YTW315 was monitored by subculturing into 9K liquid medium containing varied concentrations of CuSO4, BaSO4, FeSO4, Fe2(SO4)3, ZnSO4, CoSO4, MnSO4, Al2(SO4)3, CdSO4, PbSO4, NiSO4 and NaAsO3. The metals concentrations used were as follows: CuSO4, 20, 30, 40, 50 and 60 mmol/L; BaSO4, 3, 4, 5, 6 and 10 mmol/L; FeSO4, 250, 300, 350, 400 and 450 mmol/L; Fe2(SO4)3, 300, 400, 500, 600 and 1 000 mmol/L; ZnSO4, 50,100 and 150 mmol/L; CoSO4, 2, 3, 4, 5 and 6 mmol/L; MnSO4, 5, 10, 15, 20 and 25 mmol/L; Al2(SO4)3, 50, 100, 200, 300, 500 and 800 mmol/L; CdSO4, 0.5, 1.0, 3.0, 5.0 and 8.0 mmol/L; PbSO4, 0.5, 1.0, 1.5, 2.5, 3.5 and 5.0 mmol/L; NiSO4 , 5, 10, 15, 20 and 30 mmol/L; NaAsO3, 10, 30, 60 and 100 mmol/L. In metals tolerance measurements, bacteria growth was recorded as either positive or negative.
2.7 DNA preparation and G+C content
The DNA was extracted in accordance with the manufacturer’s instructions by DNA extraction kit (Tiangen Biotech Beijing, China), and the DNA base composition (G+C) content was determined using HPLC method[21].
2.8 PCR of 16S rRNA and phylogenetic analysis
The 16S rRNA genes of YTW315 were amplified by polymerase chain reaction (PCR) using the primers designed based on the previous report[22]. The PCR amplification was carried out according to the method described by DING et al[23]. PCR products of the expected size (approximately 1.5 kb) were excised from 1.0% agarose gels and purified with the purification columns (Promega), following the manufacturer’s recommendations. The PCR products were ligated to the pGM-T vector and transformed into Escherichia coli DH5α. The white colonies on the Luria-Bertani (LB) plates containing ampicillin (100 μg/mL) and X-gal (20 mg/mL) were selected and sent to Shanghai Sunbiotech Co. Ltd. for sequencing.
To construct a phylogenetic tree showing the relationship of YTW315 to other Leptospirillum species, the 16S rRNA sequences of the related reference organisms were downloaded from public databases (http://www.ncbi.nlm.nih.gov/). These were aligned with the sequence from isolate YTW315 using Clustal X 1.80. This alignment was used to make a distance matrix, and followed by a neighbor joining tree. Bootstrap analysis was carried out on 100 replicate input data sets. Phylogenetic trees were viewed using Treeview software MEGA 3.1.
2.9 PCR amplification and analysis of 16S-23S intergenic region (IR)
The protocol used for 16S-23S amplification was the same as that used for 16S rRNA gene amplification, except the annealing step which took place at 45 ℃. The primers used in amplification were G1.2: 5′-GTCGTAACAAGGTACCCG-3′ and L1.2: 5′-GCCAAGGCATCCACC-3′, which were modeled on primers designed by JENSEN et al[24].
2.10 Leaching experiments
Bioleaching tests were carried out in 250 mL flasks containing 100 mL 9K medium. The 9K basal salts medium without iron was used in the bioleaching experiments. The mineral concentration was 2% (mass (g) to volume (mL)). The inocula of L. ferriphilum culture was 10% (volume fraction), and all the experiments were carried out in triplicate. Abiotic controls were also designed by replacing the bacterial inoculum by an equal volume of related medium. Aliquots of leachate were sampled, and the concentrations of copper and iron were determined by atomic absorption spectrometer (Hatichi Z-8000) within 35 d of incubation. The lost water in the medium was supplemented with sterilized deionized water after sampling each time.
3 Results and discussion
3.1 Isolation by plating
After incubation for about 4-5 d, colonies appeared on overlaid solid media plate, and showed round, maroon and convex. The plate became yellow after 3 d, and the medium became carmine around the colonies because of oxidation of Fe(Ⅱ). Colonies can form successfully by using overlaid plate in just a few days (Fig.1(a)).
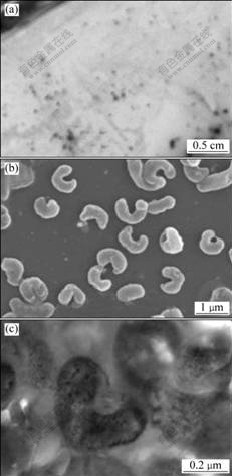
Fig.1 Photograph of strain YTW315 grown on double-layer plate (a) and its SEM image (b) and TEM image (c)
There have been numerous reports of difficulties in growing acidophilic autotroph on common solid media. At least two reasons account for the poor growth, i.e., both the soluble oligosaccharide and monosaccharide are produced by gelling of agent agar under acidic conditions; and some metabolites of the Leptospirilla affect the formation of colonies. In this study, due to the presence of Acidiphilium sp. PJH that can grow on the underlay and fully utilize the compounds in the medium, colonies of it can thus form efficiently.
3.2 Morphology and ultrastructure
Cells stained were Gram-negative and were motile, vibrio- or spiral-shaped, with a diameter of 0.3-0.5 μm and a length of 1.1-2.6 μm. As shown in Fig.1(b), cells consist mainly of vibrio-like bacteria, and seldom are spiral with up to two turns. Ultrathin sections of YTW315 show that this strain contains capsule (Fig.1(c)). Morphology results indicate that YTW315 seems to be consistent with previously described Leptospirillum species[12].
3.3 Physiological-biochemical characteristics
3.3.1 Optimal pH and temperature for growth
The effects of temperature and pH of the isolate are shown in Fig.2. The optimal temperature and pH for growth, as indicated in Fig.2, are 40 ℃ and pH 1.6, respectively. The growth is inhibited when temperature is higher than 50 ℃; and no growth is observed at pH 3.0 or above. The capability of growth at 45 ℃ strongly indicates that YTW315 is similar to Leptospirillum ferriphilum species.
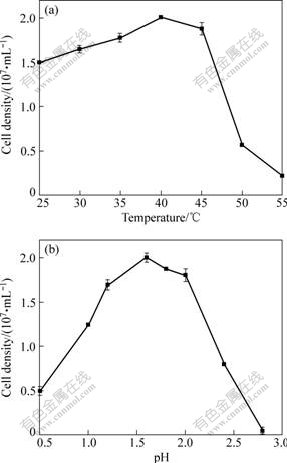
Fig.2 Effects of temperature (a) and pH (b) on growth of strain YTW315
3.3.2 Growth curve
The growth curve of strain YTW315 is shown in Fig.3. It follows the lag, logarithmic, stationary and aging phases as seen in other bacteria. The logarithmic phase occurs in 20-48 h and the number of cells reach the maximum (about 5.06×107 cell/mL) after cultivation for 52 h. The generation time and maximum specific growth rate are 9.92 h and 0.06 h-1, respectively.
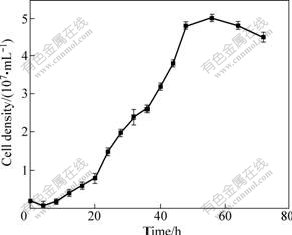
Fig.3 Growth curve of YTW315 at 40 ℃ and pH 1.6
3.3.3 Tolerance to heavy metals
As mentioned above, the isolate YTW315 was isolated from extremely acidic environments containing kinds of heavy metals. This suggests the isolate would be tolerant to heavy metals. The minimal inhibition concentrations (MICs) of some heavy metals for strain YTW315 were determined and the results are shown in Fig.4. It is shown that strain YTW315 has strong tolerance to Al(Ⅲ) and Fe(Ⅲ), and a higher tolerance to As(Ⅴ) with the concentration 50 mmol/L. Such heavy metals tolerance confers strain YTW315 a special advantage in bioleaching.
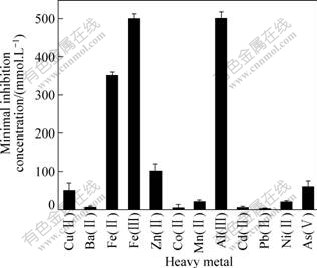
Fig.4 Tolerance of isolate YTW315 to some heavy metals
3.3.4 DNA base composition
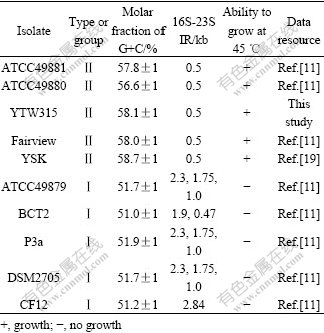
It was reported that only group Ⅱ Leptospirilla were capable of growing in the temperature range of 35-45 ℃ or even higher temperature, due to the fact that Group Ⅱ Leptospirilla have a guanine plus cytosine (G+C) content from 55% to 58%[11]. Chromosomal base analysis shows that isolate YTW315 has a (G+C) content of 58.1%, which may mainly confer it higher optimum growth temperature than L. ferrooxidans. This thus suggests that the isolate is a group Ⅱ Leptospirillum rather than L. ferrooxidans.
3.4 Phylogenetic analysis of 16S rRNA and 16S-23S rRNA
The 16S rRNA as well as 16S-23S rRNA was amplified and the PCR amplification product was detected by 1.0% agarose gel electrophoresis. The result is shown in Fig.5. The length of 16S rRNA is about 1.5 kb and is sequenced sequentially. It was submitted to the GenBank and the accession number EU733647 was obtained. Based on the homology of 16S rRNA, the phylogenetic development tree was built, as shown in Fig.6. The sequences were divided into two groups: Group Ⅰ, L. ferrooxidans and Group Ⅱ, L. ferriphilum. Isolate YTW315 was clustered into Group Ⅱ, L. ferriphilum and possessed 99.8% sequence similarity with the typical L. ferriphilum strains Fairview and ATCC49881. Also as shown in Table 2, 16S-23S rRNA gene spacer regions of YTW315 (Fig.5(b), 500 bp) further demonstrate that strain YTW315 belongs to the species Leptospirillum ferriphilum. In Fig.5, Acidithiobacillus ferrooxidans is used as a member of outgroups to root the tree, and the database accession numbers of the gene sequences used are given in parentheses. Bootstrap values obtained with 1 000 bootstrap re-samplings are given at branching points of interest. The scale bar represents 10 nucleotide substitutions per 1 000 nucleotides.

Fig.5 Agarose gel electrophoresis of PCR-amplified 16S rRNA(a) and 16S-23S intergenic region (b): 1—Marker (1kb, ferment molecular biochemicals); 2—16S rRNA of YTW315; 3, 3′—Blank control; 4—DNA marker; 5—16S-23S rRNA of YTW315
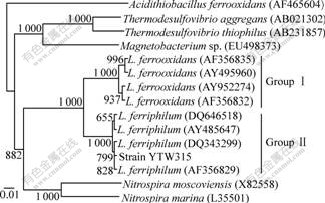
Fig.6 Distance-matrix tree showing phylogenetic affiliations of isolate YTW315, and related reference organisms based on 16S rRNA sequences
Table 2 Comparison in some molecular characteristics of YTW315 and members of genus Leptospirillum
3.5 Leaching results
3.5.1 Bioleaching of pyrite by L. ferriphilum YTW315
The bioleaching results of pyrite by YTW315 at 40 ℃ are shown in Fig.7. In the whole process, total iron extraction continuously increased. From 10 to 25 d, leaching rate of iron increased significantly and finally reached 95.31% after 30 d. Almost no soluble iron was detected in un-inoculated controls all the time. The data indicate that L. ferriphilum YTW315 has great capacity of leaching pyrite, and this result is consistent with previous reports[19].
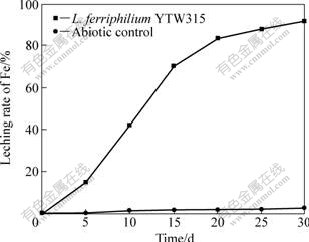
Fig.7 Leaching rate of pyrite leached by L. ferriphilum YTW315 at pH 1.6 and 40 ℃
3.5.2 Bioleaching of chalcopyrite by L. ferriphilum YTW 315
As shown in Fig.8, the extraction rate of copper increased with the bioleaching time, and was up to 44.56% after 30 d. In first 20 d, the leaching rate kept high until copper concentration became an inhibitor of the growth of L. ferriphilum YTW315. The tendency of chalcopyrite oxidation tended to be stable for L. ferriphilum YTW315 as well as the un-incubated controls after 20 d. With the continuance of bioleaching, jarosite precipitation resulted in the decrease of soluble iron in the leaching solution, and formed a passivation layer on the mineral surface[25]. The passivation layer may strongly inhibit ferric iron reduction and thus decrease the copper leaching rate. In addition, more and more copper ions accumulated in leaching process may embarrass the growth of L. ferriphilum, consequently, affect the extraction rate of copper.
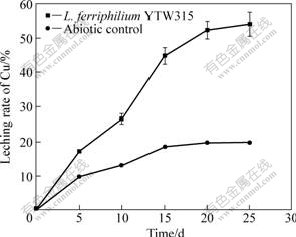
Fig.8 Leaching rate of chalcopyrite leached by L. ferriphilum YTW315 at pH 1.6 and 40 ℃
4 Conclusions
1) A new strain L. ferriphilum YTW315 is successfully isolated by overlay agar solid medium. This medium is more efficient for isolation of L. ferriphilum compared with other methods. In this medium, it is presumed that Acidiphilium sp. PJH facilitates the formation of L. ferriphilum colonies.
2) The cells of YTW315 are Gram-negative and are motile, vibrio- or spiral-shaped, with a diameter 0.3-0.5 μm and a length of 1.1-2.6 μm. It has a guanine plus cytosine (G+C) content of 58.1% and exhibits similarity of 99.8% to L. ferriphilum strain Fairview and ATCC 49881.
3) Physiological investigation indicates that L. ferriphilum YTW315 is a strict chemolithoautotroph, metabolizing ferrous iron and pyrite. The optimal temperature of L. ferriphilum YTW315 is 40 ℃ and the optimal pH is 1.6 for growth. The strain has high tolerance to As(Ⅴ) with the concentration 50 mmol/L.
4) The leaching rates of iron and copper by L. ferriphilum YTW315 are 95.31% and 44.56% after 30 d, respectively, and the results reveal L. ferriphilum YTW315 is very useful in bioleaching of metal sulfides ores.
References
[1] BRIERLEY C L. How will biomining be applied in future [J]. Transactions of Nonferrous Metals Society of China, 2008, 18(6): 1302-1310.
[2] POULIN R, LAWRENCE R W. Economic and environmental niches of biohydrometallurgy [J]. Minerals Engineering, 1996, 9(8): 799-810.
[3] ROHWERDER T, GEHRKE T, KINZLER K. Bioleaching review A: Progress in bioleaching fundamentals and mechanisms of bacterial sulfide oxidation [J]. Applied and Environmental Microbiology, 2003, 63(3): 239-248.
[4] JOHNSON D B. Biodiversity and interactions of acidophiles: Key to understanding and optimizing microbial processing of ores and concentrates [J]. Transactions of Nonferrous Metals Society of China, 2008, 18(6): 1367-1373.
[5] NAOKO O, MARIEKIE G, HALLBERG B. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred-tank bioleaching operation [J]. Applied and Environmental Microbiology, 2003, 69(4): 1936-1943.
[6] WULF-DURAND P DE, BRYANT L J, SLY L I. PCR-mediated detection of acidophilic, bioleaching-associated bacteria [J]. Applied and Environmental Microbiology, 1997, 63(7): 2944-2948.
[7] TIAN Jian, WU Ning-feng, LI Jiang, LIU Ya-jie. Nickel-resistant determinant from Leptospirillum ferriphilum [J]. Applied and Environmental Microbiology, 2007, 73(7): 2364-2368.
[8] RAWLINGS D E, TRIBUTSCH H, HANSFORD G S. Reasons why ‘Leptospirillum’-like species rather than Thiobacillus ferrooxidans are the dominant iron-oxidizing bacteria in many commercial processes for biooxidation of pyrite and related ores [J]. Microbiology, 1999, 145(3): 5-13.
[9] SCHRENK M O, EDWARDS K J, GOODMAN R M, HAMERS R J. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: Implications for generation of acid mine drainage [J]. Science, 1998, 279(5356): 1519-1522.
[10] DOPSON M, LINDSTROM E B. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite [J]. Microbial Ecology, 2004, 48(1): 19-28.
[11] CORAM N J, RAWLINGS D E. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40 ℃ [J]. Applied and Environmental Microbiology, 2002, 68(2): 838-845.
[12] HIPPE H. Leptospirillum gen. nov. (ex Markosyan 1972), nom. rev., including Leptospirillum ferrooxidans sp. nov. (ex Markosyan 1972) nom. rev. and Leptospirillum thermoferrooxidans sp. nov. (Golovacheva et al. 1992) [J]. International Journal of Systematic Bacteriology, 2000, 50: 501-503.
[13] BOND P L, BANFIELD J F. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments [J]. Microbial Ecology, 2001, 41(2): 149-161.
[14] TYSON G W, CHAPMAN J, HUGENHOLTZ P, ALLEN E E. Community structure and metabolism through reconstruction of microbial genomes from the environment [J]. Nature, 2004, 428: 37-43.
[15] BOND P L, SMRIGA S P, BANFIELD J F. Phylogeny of microorganisms populating a thick, subaerial predominantly lithotrophic biofilm at an extreme acid mine drainage site [J]. Applied and Environmental Microbiology, 2000, 66(9): 3842-3849.
[16] TYSON G W, LO I, BAKER B J, ALLEN E E, HUGENHOLTZ P, BANFIELD J F. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community [J]. Applied and Environmental Microbiology, 2005, 71(10): 6319-6324.
[17] JOHNSON D B, MCGINNESS S. A highly efficient and universal solid medium for growing mesophilic and moderately thermophilic, iron-oxidizing, acidophilic bacteria [J]. Journal of Microbial Methods, 1991, 13(2): 113-122.
[18] LIU Ying, QI Fang-jun, LIU Xiang-mei. Phylogenetic analysis for 16S rDNA sequence of the vibrio shaped chemoautolithotrophic iron-oxidizing bacterium ML-04 [J]. Journal of Shandong University, 2004, 39(5): 112-115. (in Chinese)
[19] GAO Jian, ZHANG Cheng-gui, WU Xue-ling, WANG Hai-hua, QIU Guan-zhou. Isolation and identification of a strain of Leptospirillum ferriphilum from an extreme acid mine drainage site [J]. Annals of Microbiology, 2007, 57(2): 171-176.
[20] PENG Juan-hua, ZHANG Rui-yong, ZHANG Qian, ZHANG Li-min, ZHOU Hong-bo. Screening and characterization of Acidiphilium sp. PJH and its role in bioleaching [J]. Transactions of Nonferrous Metals Society of China, 2008, 18(6): 1443-1449.
[21] MESBAH M, PREMACHANDRAN U, WHITMAN B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography [J]. International Journal of Systematic Bacteriology, 1989, 39(2): 159-167.
[22] XIA Jin-lan, PENG An-an, HE Huan, YANG Yu, LIU Xue-duan, GIU Guan-zhou. A new strain Acidithiobacillus albertensis BY-05 for bioleaching of metal sulfides ores [J]. Transactions of Nonferrous Metals Society of China, 2007, 17(1): 168-175.
[23] DING Jian-nan, HE Huan, ZHANG Cheng-gui, YU Yi-zun, QIU Guan-zhou. Isolation and characterization of YNTC-1, a novel Alicyclobacillus sendaiensis strain [J]. Journal of Central South University of Technology, 2008, 15(4): 508-514.
[24] JENSEN M A, WEBSTER J A, STRAUS N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms [J]. Applied and Environmental Microbiology, 1993, 59(4): 945-952.
[25] STOTT M B, WATLING H R, FRANZMANN P D, SUTTON D. The role of iron-hydroxy precipitates in the passivation of chalcopyrite during bioleaching [J]. Minerals Engineering, 2000, 13(1/10): 1117-1127.
Foundation item: Projects(50621063, 50674101) supported by the National Natural Science Foundation of China; Project (2004CB619204) supported by the National Basic Research Program of China
Corresponding author: XIA Jin-lan; Tel: +86-731-88836944; E-mail: jlxia@mail.csu.edu.cn
DOI: 10.1016/S1003-6326(09)60110-2
(Edited by YANG Hua)


