J. Cent. South Univ. Technol. (2007)03-0324-06
DOI: 10.1007/s11771-007-0064-6

Isolation and characterization of organic-sulfur degradation bacterial strain
YANG Yu(杨 宇)1, 2, DIAO Meng-xue(刁梦雪)1, SHI Wu-yang(师舞阳)1, LI Li(历 丽)1,
DAI Qin-yun(代沁芸)1, QIU Guan-zhou(邱冠周)1, 2
(1. School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China;
2. Key Laboratory of Biometallurgy of Ministry of Education, Changsha 410083, China)
Abstract: A bacterial strain that was capable of degrading organic sulfur (dibenzothiophene) was isolated by enrichment techniques from the petroleum-contaminated soil collected from Zhongyuan Oil Field. The strain is named ZYX and is gram-positive. This strain undergoes bacilus-coccus morphological change, and forms yellow-pigment glossy circular colonies with 1.5 mm in diameter on average after 2 d incubation on Luria-Bertani(LB) plates. The full-length of 16S rDNA sequence of strain ZYX was determined and analyzed. Strain ZYX is found most relative with the genus of Arthrobacter. The similarity values between ZYX and Arthrobacter sp. P2 is 99.53%. The main morphological, biochemical and physiological features of strain ZYX accord with those of Arthrobacter. It is found that the optimal initial pH for growth is about 7.0, and the optimal concentration of dibenzothiophene(DBT) for growth is 0. 10 g/L. Additionally, the results show that the best carbon source and nitrogen source are glycerol and glutamine, respectively.
Key words: Arthrobacter; 16S rDNA; dibenzothiophene; organic-sulfur degradation
1 Introduction
Coal has been used as the predominant energy resource for power generation for many years in China. Fossil-fired power plants use coal as the major fuel to generate electricity. The SO2 released from power plants after direct burning will be a harmful contamination to the environment[1]. In the past couple of decades, growing concern over environmental interests has posed a problem for the coal industry in that environmental problems are caused by coal processing and combustion. More and more efforts have been committed to develop clean coal technologies by scientists and engineers. Interdisciplinary research is centered on the basic and innovative studies for the use of coal in an efficient and environmentally acceptable manner[2]. The sulfur contents can be divided into inorganic sulfur and organic sulfur. The pyretic sulfur FeS2 is the major substance of inorganic sulfur. The organic sulfur structures in coals are mainly components of the macromolecular structures of the coal and are not readily separated and analysed without destruction of the macromolecular network[3]. The structures of the organic sulfur in the coal include mercaptans, sulfides, disulfides and thiophenes. Usually, biodesulfurization is studied with model compounds, such as dibenzothiophene (DBT)[4].
The desulfurization ways can be classified into physical way, chemical way and biological way[5]. Microorganisms have developed varieties of essential biochemistry strategies to deal with sulfur contamination[6]. Compared with other methods, biodesulfurization can remove organic sulfur and dispenses at the high temperature and high pressure. Additionally, it is relatively economical. For all these reasons, more scientists focus their study on this novel method[7]. Several bacterial strains belonging to the genera Rhodococcus, Bacillus, Corynebacterium and Arthrobacter can degrade DBT through sulfur-specific pathway, in which the specific cleavage of carbon-sulfur bonds of DBT occurs to yield 2-hydroxybiphenyl (2-HBP) without degradation of the organic structure, meaning that they would accomplish the desulfurization of coal and petroleum without a loss of heating value[8].
In this study, a bacterial strain was isolated and characterized, which was named ZYX. Subsequently, the growth characterization of ZYX was studied, which contains pH, carbon sources, nitrogen sources, sulfur sources and different concentrations of DBT.
2 Materials and method
2.1 Microorganism source
Bacterial strain came from the petroleum- contaminated soil, which was collected from Zhongyuan Oil Field.
2.2 Chemicals
Dibenzothiophene (DBT) and 2-hydroxybiphenyl (2-HBP) were purchased from Fluka. L-valine and L-histidine were purchased from Sino-American Biotechnology Company. L-phenylalanine and L-serine were purchased from Shanghai Sangon Biological Engineering & Technology and Service Corporation, China. L-glutamine was purchased from Ding Guo Biotechnology Corporation (Beijing, China). KH2PO4, NH4Cl, Na2HPO4·12H2O, MgCl2·6H2O, glycerol, saccharose and tri-sodium citrate were analytically pure.
2.3 Media
The basal salt medium(BSM) was as follows (g/L)[9]: KH2PO4 2.44, Na2HPO4·12H2O 14.03, MgCl2·6H2O 0.36, NH4Cl 2.00, MnCl2·4H2O 0.004, FeCl3·6H2O 0.001, CaCl2 0.001, glycerol 1.62. It was sterilized at 121 ℃ for 20 min. The basal salt DBT medium (BSDM) was formed by adding 0.1 g/L DBT into BSM as the sole sulfur source. The basal salt coal medium (BSCM) was formed by adding coal powder into BSM as the sole sulfur source. The solid medium was BSDM and 1.5% agar.
The bacterium was cultivated on four kinds of solid plates[10] as follows: LB (Luria-Bertani) plate, Gause No.1 medium, Czapek medium and starch medium.
2.4 Enrichment and isolation
10 g sample soil collected from the oil field was uniformly dissolved with 500 mL distilled water. Sequentially the mixture was standed for 5 min. 5 mL supernatant was inoculated into 100 mL BSDM in a 250 mL Erlenmeyer flask. Enrichment cultures were incubated at 30 ℃ with shaking for 5 d at 170 r/min. The enrichment cultures obtained from the above step were diluted for 10-3, 10-4 and 10-5. The diluted solutions of enrichment cultures were spread on the solid medium and incubated in the incubator at 30 ℃ for 2-3 d. The colonies were picked up and inoculated into 100 mL BSDM. Then the cultures were incubated with shaking at 30 ℃ and 170 r/min. After successive transfers to new plates, individual and distinguishable colonies were selected for identification and further experiments.
2.5 Analytical methods
2.5.1 Cell amount
Population of the isolate was counted by haemacytometer under optical microscope for enumeration generation[11].
2.5.2 Morphological characterization
The morphology and rod-coccus growth cycle of the isolate were observed by the optical microscope. Gram staining was done to purify culture isolate according to routine method. Scanning electron micrographs were taken for the isolated strain. The morphological characteristics of the isolate were assessed. Sample preparation was as follows. The fermentation liquor cultured for 72 h was gathered in a 2.0 mL EP (Eppendorf) tube and spun at 15 kr/min for 10 min. The flow-through was discarded. The bacteria were resuspended and washed with TE (Tris-EDTA: 10 mmol/L Tris-HCl, pH 7.6; 1 mmol/L EDTA, pH 8.0). The sample was spun for another 10 min. The two steps above were repeated for 3 or 4 times to get enough bacteria. The sample was fixed in glutaraldehyde. The scanning electron micrographs were taken in College of Life Science, Hunan Normal University, China.
2.5.3 16S rDNA characterization
DNA extraction of the isolate followed the directions of EZ-10 Spin Column Genomic DNA Minipreps Kit (Protocol-BS423&BS424, Bio Basic Inc.). The 16S rRNA genes were amplified from the genomic DNA using universal bacterial primers F27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and R1492 (5′-GGTTACCTTGTTACGACTT-3′). The conditions used for ploymerase chain reaction(PCR) were: 10×dNTPs (2 mmol/L each) 5 μL, 10×PCR buffer 5μL, target DNA (0.1 μg/μL) 2 μL, enzyme mixture (volume ratio of Taq DNA polymerase to Pfu DNA polymerase 1:1) 1 μL, primers 10 pmol and the reaction mixture made to a final volume 50 μL with deionized water. Thermal cycling was as follows: pre-denaturation at 94℃ for 5 min, followed by denaturation at 94 ℃ for 1 min, annealing at 55 ℃ for 1 min and extension at 72 ℃ for 3 min of 30 cycles.
The PCR products were purified by E.Z.N.ATM gel extraction Kit (OMEGA.USA), according to the instructions of the manufacture. The purified products were cloned into PCR2.1 vector using TA cloning kit (Invitrogen, San Diego, CA, USA). Clones were sequenced by Sunbiotech Company (Beijing, China). The 16S rDNA sequences were analyzed and aligned with other sequences deposited in Genbank by ClustalX[12] to construct the phylogenetic tree. The sequence data were compared with 16S rRNAs sequence deposited in public databases by using the BLAST (Basic Local Alignment Search Tool) search program.
2.5.4 Growth characterization
The growth characterization of ZYX was studied, which contains pH, carbon sources, nitrogen sources, sulfur sources and different concentrations of DBT.
3 Results and discussion
3.1 Morphological characterization
The shapes of colonies on agar plates of different media are described in Table 1. The strain ZYX on the LB plate is buff round colony, edge integrated, wet, glossy and convex. The colonies are measured as 1.5 mm in diameter on average after a 2 d incubation (Fig.1). The buff colonies turn to full yellow with the culture time. The strain is gram-positive but easily faded. Using SEM we determined the cellular dimensions: (0.7-0.9) μm×(2.0-3.0) μm. Strain ZYX is in irregular coryneform and arranged in ‘V’-shape (Fig.2). During the growth cycle, almost microbes are bacilus within initial 5 h, however, most bacilus break into cocci after 20 h.
Table 1 Colony characteristics on agar plates of different media

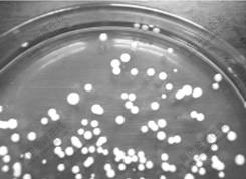
Fig.1 Photograph of strain ZYX on LB plate

Fig.2 Scanning electron micrograph of strain ZYX
The strain was primarily characterized by the optical microscopy, gram-staining, SEM method, colony shape on the culture plates, “Bergey’s manual of systematic bacteriology (Eighth edition)”[13] and “Common identification technique of normal bacteria”[14]. It is demonstrated that strain ZYX belongs to Arthrobacter.
3.2 16S rDNA gene sequencing and analysis
The length of the sequence is 1 481 bp. The strain sequence was compared with 16S rRNA sequence deposited in public databases by using BLAST program of National Center for Biotechnology Information (NCBI). The results show that strain ZYX shares 99.53% identities with the type strain of Arthrobacter sp. P2 (DQ288888.1, Genbank) for 16S rRNA gene (Table 2). The Genbank accession number for its 16S rRNA gene sequence is DQ648530.
Table 2 Closest bacteria to strain ZYX according to 16S rDNA sequence

The 16S rDNA sequences were analyzed and aligned with other sequences deposited in Genbank by ClustalX program to construct the phylogenetic tree. Phylogenetic analysis was carried out using the sequence of strain ZYX, the sequences within Escherichia coli genus and other close relatives from the Genbank. A maximum likelihood tree generated by ClustalX is shown in Fig.3. Strain ZYX is found to cluster tightly with the type strain Arthrobacter sp. P2. Moreover, strain ZYX shares 99.13% and 99.18% identities respectively with Arthrobacter aurescens and Arthrobacter nitroguajacolicus that have been determined for specific species. The strain ZYX is on the same ramification with Arthrobacter sp. P2 and Arthrobacter sp. 68 m. It is dubious which species strain ZYX belongs to, because the two strains mentioned above are not determined for specific species. Fortunately, we are sure that strain ZYX belongs to Arthrobacter genus. Thus there are still further studies that should be carried out.

Fig.3 Phylogenetic tree based on 16S rRNA sequence analysis of members of genus Arthrobacter showing position of Arthrobacter strain ZYX(Accession numbers of reference sequences used in phylogenetic analysis are as follows: Escherichia coli, E05165; A. protophormiae, AY577525; A. oxydans, AJ243423; A. sp.P2, DQ288888; A. sp. 68m, AJ853464; A. aurescens, DQ016989; A. nitroguajacolicus, AJ512504; A. sp. KA1-1, AJ785760; A. ilicis, X83407; A. nicotinovorans, AY833098; A. histidinolovorans, X83406)
3.3 Growth characterization
3.3.1 Effect of different initial pH on strain growth
The result is shown in Fig.4. The bacterial amount is little in the acid cultures (pH<5.0) and the alkaline cultures (pH>10.0). The bacterial amount of strain ZYX is the largest when initial pH is between 6.0 and 7.0. Furthermore, it is demonstrated that strain ZYX can survive in a wide range of pH value and it prefers the neutral environment.

Fig.4 Effects of different pH values on growth of strain ZYX
3.3.2 Growth curve of strain ZYX
The results obtained during the growth of the strain ZYX are shown in Fig.5. The lag phase is from 0 to 22 h; the log phase is from 22 to 36 h, the stationary phase is from 36 to 60 h; and the death phase begins from 60 h. It is shown that the stationary phase is relatively short because of the consumption of energy sources.

Fig.5 Growth curve of strain ZYX
3.3.3 Influence of different carbon sources
Five kinds of carbon sources were investigated: glycerol, glucose, saccharose, amidulin and tri-sodium citrate. The initial pH was 7.0 and the inoculum amount was 2 mL. Experimental results are shown in Fig.6. As

Fig.6 Effect of different energy sources on strain growth
(a) Carbon source; (b) Nitrogen source; (c) Sulfur source
(S represents sublimed sulfur)
shown in Fig.6(a), the strain can utilize all these carbon sources; and the best result is obtained when glycerol is employed as carbon source. In this case, growth is faster than that observed in the other carbon sources. Amidulin cannot be used as the sole carbon source by strain ZYX.
3.3.4 Influence of different nitrogen sources
The source of nitrogen usually used for the growth of the desulfurizing microorganisms is ammonium chloride at a concentration of 2 g/L[15]. Therefore, 2 g/L of this nitrogen source was chosen in present work. As shown in Fig.6(b), L-glutamine has a clear influence. The cell amount is higher when L-glutamine is used as the only nitrogen source. Additionally, the strain can hardly utilize L-histidine (heterocyclic amino acid) as sole nitrogen source. The cell amount is higher when NH4Cl is employed. In conclusion, the strain ZYX grows better in organic nitrogen sources than inorganic ones. In addition, it is necessary to consider that these amino acids can be used as both carbon and nitrogen sources of growth, allowing possibilities for comparison of two kinds of nitrogen source carried out in the experiments.
3.3.5 Influence of different sulfur sources
In order to study the influence of sulfur sources, the growth of strain ZYX using different sulfur sources, including sublimed sulfur, sodium hyposulfite (Na2S2O3), ferrous sulfate (FeSO4), benzenesulfonic acid (C6H5SO3H) and DBT, was carried out at a concentration of 0.01 g/L, and an initial pH of 7.0. In Fig.6(c), the results obtained during the growth of strain ZYX employing different sulfur sources are shown. Clear differences can be observed: the cell amount is the largest when sublimed sulfur is used as sulfur source rather than DBT. The strain can utilize both organic and inorganic sulfur. Moreover, it grows better in the media containing inorganic sulfur. A brief experiment was done to determine whether the strain could propagate in the culture medium without sulfur source DBT.
3.4 Effect of initial DBT concentration on growth of
strain ZYX
In this experiment, different concentrations of DBT (0.01, 0.05, 0.10 and 0.20 g/L) were studied (Fig.7) in order to determine the influence of initial concentration of DBT on the strain growth. Clear differences can be observed: the optimal concentration of DBT is 0.10 g/L. Higher DBT concentration can lead to inhibition, while lower DBT concentration cannot meet the need for strain growth.
3.5 Growth of strain ZYX in media containing coal
The strain ZYX was inoculated into cultures containing coal powder as the only sulfur source. The content of coal powder in the samples of A, B, C were 0.02, 0.05 and 0.10 g/mL respectively. In Fig.8, the bacterial amount in sample C is the smallest among the three samples after 6 h incubation, while that in sample A is the largest. After 24 h incubation, the bacterial amount in sample B becomes the largest.

Fig.7 Effect of initial DBT concentration on growth of strain ZYX

Fig.8 Growth curve of strain ZYX in culture medium containing coal
4 Conclusions
1) The strain is gram-positive, but easily faded. The colonies on LB plates are buff, circular, smooth, wet, convex, entire margins and slightly glistening. The scanning electron micrographs indicate that this strain is in irregular coryneform. The cellular dimensions are (0.7-0.9) μm×(2.0-3.0) μm. Some cells are arranged at an angle in a “V” formation. The fermentation liquor is offwhite within a 3 d shaking cultivation and becomes fully yellow after 3 d yielding flocculent depositions. The morphological change is obvious. During the stable phase cocci predominates.
2) The result of 16S rDNA analysis demonstrates that this strain named ZYX belongs to Arthrobacter genus and shares 99.53% identities with Arthrobacter sp. P2.
3) The optimal growth condition for the isolate is as follows: pH 7.0, glycerol as best carbon source, and L-glutamine as best nitrogen source. This strain can use glycerol, saccharose, glucose and tri-sodium citrate as carbon source. Amidulin cannot be used. Both organic and inorganic sulfur can be utilized. The optimal DBT concentration for strain growth is 0.10 g/L. Higher DBT concentration can lead to inhibition, while lower DBT concentration cannot meet the need for strain growth. Strain ZYX could utilize organic sulfur in coal as the sole sulfur source to survive in BSCM. The optimal coal powder content is 5×10-2 g/mL.
References
[1] WANG Zhi-xuan, PAN Li, PENG Jun. Analysis of current status, cost and policies of power sector SO2 emission control[J]. Research of Environmental Sciences, 2005, 18(4): 11-20. (in Chinese)
[2] LEI Fang-ming. Review of technology on fuel coal sulphur removal[J]. Energy Conservation, 2002, 243(10): 16-18. (in Chinese)
[3] QIU Jian-hui, DI Jin-shen, LI Ying-jie. Progress of biodesulfurization[J]. Acta Microbiologica Sinica, 2001, 41(5): 650-653. (in Chinese)
[4] DEL OLMO C H, SANTOS V E, ALCON A, et al. Production of a Rhodococcus erythropolis IGTS8 biocatalyst for DBT biodesulfurization: Influence of operational conditions[J]. Biochemical Engineering Journal, 2005, 22: 229-237.
[5] SUN Li-mei, SHAN Zhong-jian. Research progress of coal desulfurization at host combustion in domestic and abroad[J]. Clean Coal Technology, 2005, 11: 55-59. (in Chinese)
[6] ZHOU Zhi-fu, WEI De-zhou, WANG Ying-min, et al. The progress of the research on microbial pretreatment and floatation desulphurization in coal[J]. Industrial Safety and Environmental Protection, 2002, 28(2): 3-7. (in Chinese)
[7] CHENG Sheng-gao, WANG Yan-lin. Stored-up status of organic sulfur in high sulfuric coal and desulfurized mechanism with microbe[J]. China Environmental Protection Industry, 1996, 12: 34-35. (in Chinese)
[8] DAHLBERG M D, ROHRER R L, FAUTH D J, et al. Biodesulfurization of dibenzothiophene sulfone by Arthrobacter sp. and studies with oxidized Illinois[J]. 1993, 72: 1645-1649.
[9] HOU Zhong-xuan, LIU Hui-zhou, LUO Ming-fang, et al. Isolation and characterization of a desulfurization bacterium[J]. Science in China (B), 2002, 32(5): 397-405. (in Chinese)
[10] SHEN Ping, FAN Xiu-rong, LI Guang-wu, et al. Microbiological Experiments[M]. The Third Edition. Beijing: Higher Education Press, 1999.
[11] XU Wei-chang. Application of Biotechnology in Nuclear Industry[M]. Hunan: National University of Defense Technology Press, 2002. (in Chinese)
[12] THOMPSON J D, GIBSON T J, PLEWNIAK F, et al. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Research, 1997, 24: 4876-4882.
[13] KEDDIE R M, COLLINS D, JONES D, SNEATH P H A, MAIR N S, SHARPE M E, Holt J G (eds). (1986a) Genus Arthrobacter. Bergey’s Manual of Systematic Bacteriology(Vol.2)[M]. Baltimore: Williams & Wilkins, 1986: 1288-1301.
[14] Microorganism Institute, The Chinese Academy of Sciences. Common Identification Technique of Normal Bacteria[M]. Beijing: Science Press, 1984. (in Chinese)
[15] CAROLINA H D, ALMUDENA A, VICTORIA E S, et al. Modeling the production of a Rhodococcus erythropolis IGTS8 biocatalyst for DBT biodesulfurization: Influence of media composition[J]. Enzyme and Microbial Technology, 2005, 37: 157-166.
(Edited by YANG Bing)
Foundation item: Project(50621063) supported by the National Natural Science Foundation of China; Project(05JJ30023) supportecd by Natural Science Foundation of Hunan Province, China
Received date: 2006-06-28; Accepted date: 2006-10-27
Corresponding author: QIU Guan-zhou, Professor ; Tel: +86-731-8877216; E-mail: biocsu@126.com