J. Cent. South Univ. Technol. (2009) 16: 0053-0060
DOI: 10.1007/s11771-009-0009-3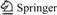
Identification and fermentation optimization of protopectinase-overproducing strain Aspergillus niger CD-01 for pectin production
XIA Jin-lan(夏金兰)1, 2, MENG Hao(孟 浩)2, WANG Run-min(王润民)2,
ZHANG Cheng-gui(张成桂)2, XIONG Jing(熊 晶)2, NIE Zhen-yuan(聂珍媛)1, 2, QIU Guan-zhou(邱冠周)1, 2
(1. Key Laboratory of Biometallurgy of Ministry of Education, Central South University,Changsha 410083, China;
2. School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China)
________________________________________________________________________
Abstract: In order to solve the citrus peel resource waste problem and minimize the drawbacks of chemical extraction of pectin, a protopectinase-overproducing strain CD-01 for pectin production was isolated from a pit soil dumped with perished orange in Changde City, Hunan Province of China. The strain CD-01 had the same morphology and 28S rRNA gene sequence (FJ184995) as that of Aspergillus niger (ATCC 64028). It was thus identified and named as Aspergillus niger CD-01. The fermentation condition was optimized based on L9(34) orthogonal experimental design and the variances analyses. The results show that the optimal condition for producing pectin is as follows: time 36 h, temperature 35 ℃, pH 5, and urea as the nitrogen source. Under this condition, the pectin yield can reach up to 24.5%. This shows a great potential of Aspergillus niger CD-01 in pectin extraction from citrus.
Key words: Aspergilllus niger; pectin production; protopectinase; citrus peel; fermentation optimization
________________________________________________________________________
1 Introduction
Pectin [1] is widespread in fruits, roots and leaves of plants as the forms of protopectin [2], soluble pectin and pectic acid, which is a very important food additive, and has been lacked extremely in Chinese market with price of about 17 000 US$ per ton.
Citrus peel, a by-product of the citrus processing industry and typically with about 30% (on dry peel), is one of the main source of pectin in industry.
Most commercial pectin has still been produced by chemical extraction in China. The process of chemical extraction of pectin is energy-consuming and produces a large amount of industrial wastes. A fermentation method, however, can minimize such drawbacks and produce pectin with higher quality. In this way, TAKUO and MINORU [3] isolated different microorganisms able to produce protopectinase to extract pectin from citrus peel, and the pectin yield could up to 13.3% (on dry peel) [4]. After that, protopectinase was considered as pectin-releasing or pectin-sobilizing enzyme. In 1999, ESQUIVEL et al [5] used acidic protopectinase that was isolated from culture of Aspergillus kawachii IFO 4308 to extract pectin from lemon peel, and the pectin yield was up to 17.4% (on dry peel). Production of pectin from citrus peel by fermentation had been adopted in industry in foreign countries, while it has not happened yet to date in China. And the pectin yield of fermentation is much lower than the yield of other physical and chemical methods until now [6]. It will be obviously meaningful to screen a better strain and optimize the fermentation condition.
Protopectinases are a variety of enzymes that can solubilize pectin from the insoluble plant protopectin and play the major role in the fermentation process, and pectinase [7] is a variety of enzymes that degrade pectin during fermentation. It means that higher activity of protopectinases and lower activity of pectinases during fermentation may lead to higher pectin yield. In this paper, the activities of these two kinds of enzymes were monitored to optimize the fermentation condition for producing pectin.
2 Experimental
2.1 Reagents
Pectin (analytical reagent) was product of Sigma; protopectin was self-made [8]; X-gal (5-bromo- 4-chloro-3-indolyl-β-D-galactoside) was product of Amresco; IPTG (Isopropyl-β-D-thiogalactopyranoside) was product of Merck; Ampicillin was product of Amresco; D-(+)galacturonic acid monohydrate (biological reagent) and carbozole (chemical purity) were products of Sinopharm Chemical Reagent Co. Ltd. CTAB (cetyltrimethyl ammonium bromide) was used for extraction of genomic DNA, sterilized 6.25% (mass fraction) of dry citrus peel was used for culture medium for pectin yield assay, and Cz`apek-Dox medium with addition of 0.5% (mass fraction) of pectin culture medium was used for enzyme activity assay, and Gel Recovery Kit (E.Z.N.A?) was used for the polymerase chain reaction (PCR) product retrieve. All other reagents were analytical reagent and made in China, and sterilized water was used.
2.2 Source of strains
The sample was collected from the soil 10 cm below the surface of the soil in a hole that dumped perish orange in Changde City, Hunan Province, China. Citrus peel powder was used as the carbon source for the origin and the secondary screenings, in which the production of pectin and the enzyme activity of protopectinase were monitored and used as the guideline, and the higher the production and the enzyme activity were, the better the strain was. A protopectinase-overproducing strain was screened, whose production of pectin was the highest among the strains.
2.3 Morphology observation
Two grooves less than 1.5 cm in width were slited on the agar plate with a sterilized knife. The spore of the strain was inoculated on the grooves, and on which two slips were then covered. The strain was cultured at 30 ℃ for 3-4 d, and then observed by microscope with oil lens.
2.4 Amplification, sequencing and phylogenetic analysis of 28S rRNA gene
2.4.1 Genomic DNA extraction
Cells of the strain were harvested by filtration after being cultivated for 36 h. The mycelia on the filter paper were washed with some sterilized water three times, and the genomic DNA was extracted by CTAB method [9].
2.4.2 Amplification and sequencing of 28S rRNA gene
The D1/D2 domain of 28S rDNA [10] was cloned from the total DNA using PCR amplification with primers UNIR-26 (GGT CCG TGT TTC AAG ACG) and UNIF-26 (GCA TAT CAA TAA GCG GAG GAA AAG). All amplification reactions were done by using 10 ng of DNA and controls were made by replacing DNA with water. The following components were added to a sterilized 0.2 mL microcentrifuge tube. The reaction mixtures (50 ?L) contained 1.0 ?L (20 ?mol) of the primer, 1.5 ?L (1.5 units) of Taq DNA polymerase (Fermentas), 5 ?L of 10× supplied PCR buffer, 1 ?L (10 mmol/L) of each dNTP, 4 ?L (25 mmol/L) of MgCl2, and 1 ?L of DNA template. Distilled water was added to adjust the volume to 50 ?L.
The DNA amplifications were carried out under heating at 93 ℃ for 3 min, followed by 30-cycle of program (93 ℃, 30 s for denaturation; 58 ℃, 30 s for annealing; 72 ℃, 30 s for extension) and by a final DNA extension at 72 ℃ for 10 min. The amplified product was maintained at 4 ℃ after cycling [11-12].
The PCR product, which was purified using Gel Recovery Kit (E.Z.N.A?) after electrophoretic analysis, was cloned into the pGEMR-T vector and transformed into Escherichia coli TOP10F’ competent cells according to the manufacturer’s instruction. Plasmid clones were identified based on blue-white screening and grown overnight on plates with IPTG, ampicillin (100 mg/mL), and X-gal (15 mg/mL). White colonies were randomly selected and the cloned inserts were amplified with the vector primers T7 and SP6. The positive clone was sequenced by Shanghai Sangon Biological Engineering Technology & Services Co. Ltd.
2.5 Assay of enzymes activities and pectin yield
Protopectinase activity was assayed by measuring the pectic substances liberated from protopectin according to the carbazole-H2SO4 method with D- galacturonic acid monohydrate as the standard. The reaction was carried out at 37 ℃ for 1 h in a 1 mL of acetate buffer solution (100 mmol/L, pH=5.0) containing 10 mg of protopectin and 10 μL of ferment supernatant. One unit (1 U) of protopectinase activity was defined as the activity which liberates pectic substance corresponding to 1 ?mol of D-galacturonic acid under the reaction conditions mentioned above [13].
DNS (3,5-dinitrosalicylic acid) method [14] was adopted to determine the enzyme activity of pectinase, in which D-galacturonic acid monohydrate was used as the reference mono-saccharide. The reaction was carried out at 50 ℃, pH 3.5 for 10 min, the substrate was pectin. One unit of pectinase activity was defined as the activity which liberates 1 ?g of D-galacturonic acid under the reaction conditions mentioned above. The pectin yield was assayed by carbazole-H2SO4 method [15] with D- galacturonic acid monohydrate as the standard. The fermentation liquid was centrifugated for 5 min at 10 000 r/min. 150 μL of the supernatant was taken and the pectin in it was deposited by 350 μL ethanol and washed three times by 63% ethanol. The deposit was dissolved in 1 mL of 0.5 mol/L NaOH solution and the pectin in it was determined and the pectin yield was calculated [16].
2.6 Measurement of growth curve
Weighing method was adopted in the measurement of growth curve [17]. The strains were cultured at 35 ℃ in 250 mL flasks containing 100 mL culture medium. The mycelia were harvested at an interval of 4 h by filtration, and then washed with distilled water three times. After that, the mycelium was dried under the conditions of 80 ℃ for 4 h and then the mass of mycelium was measured with scale.
3 Results and discussion
3.1 Identification of strain
3.1.1 Microscopic morphology
The morphologies of the colonies, conidiophore, conidiospores and hyphae of strain CD-01 are shown in Fig.1. During the cultivation at 35 ℃, the colonies observed by nakes eyes were initially white, and quickly became black with conidial production, and the back of Petri vessel was pale yellow, and radial fissures appeared in the agar. Under optical microsope the morphologies of conditiophore, conidiospores and hyphae were also observed. Under miroscope with oil lens, the strain was observed with septate and hyaline hyphae, and 1 000- 3 000 ?m long of smooth and hyaline conidiophores, and the apex becoming darker and terminating in a globose vesicle. Conidia are brown to black, very rough, globose, and measure 5-6 ?m in diameter. These phenomena indicate that strain CD-01 could be Aspergillus niger [18-19].

Fig.1 Morphologies of colonies, conidiophore, conidiospores and hyphae of strain CD-01: (a) Colonies cultivated for 2 d; (b) Colonies cultivated for 4 d; (c) Conidiophore and conidiospores; (d) Hyphae fragment
3.1.2 Analysis of 28S rRNA gene sequence
The D1/D2 domain was amplified with primers UNIR-28 and UNIF-28. The graphs of electrophoretic bands (Fig.2) show that the PCR product is the target fragment.

Fig.2 Electrophoretic bands of PCR products (where M denotes marker, bands 1 and 2 denote PCR products of primers of D1/D2 domain, and CK denotes negative control)
The positive clone was sequenced, and 587 base pairs (bp) of D1/D2 domain of the strain were obtained, and the accession number registered in Genbank was 554FR47D015. The phylogenetic tree shown in Fig.3 summarizes the phylogenetic relationship among the Aspergillus species. It reveals that strain CD-01 is closely related to Aspergillus niger ATCC 64028 (AF454171.1) [20]. According to the microscopic morphology observation and the analysis of 28S rRNA gene sequence, the strain CD-01 could be thus identified as Aspergillus niger.
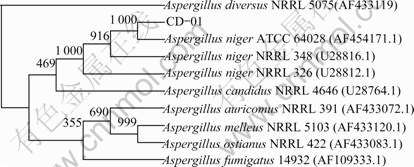
Fig.3 Phylogenetic tree derived from D1/D2 domain sequence of CD-01
3.2 Optimization of fermentation condition
3.2.1 Choice of carbon source of medium for enzyme activity assay
Citrus peel is the carbon source in the process of pectin extraction by fermentation method. However, it does not fit to use as carbon source of the medium for enzyme activity assay because of its complicated components.
Citrus peel powder, starch, glucose, pectin, sucrose, and carboxymethyl cellulose (CMC) were chosen for carbon source investigation. The effects of these carbon sources on activities of pectinase and prototinase are shown in Fig.4. It can be seen that pectin and citrus peel powder present high level of protopectinase activity, which can be explained that the protopectinase is induced by pectin and pectin contents in the citrus peel powder [21], while the pectinase activity is low, attributing to the catabolite inhibition [22]. Compared with other carbon source, the effect of pectin is similar to that of citrus peel powder. Therefore, pectin was chosen as the carbon source for enzyme activity assay.

Fig.4 Effect of carbon source on activities of pectinase and protopectinase
3.2.2 Optimization of single condition
The fermentation conditions including nitrogen source, temperature, pH value, inoculation amount, carbon source, dissolved oxygen concentration and fermentation time were firstly studied, respectively. Among these conditions, fermentation time, temperature, pH and nitrogen source showed a high level of effect on enzyme activity. Therefore, these conditions were chosen for the further study.
Supposing that Q equals to the ratio of protopectinase activity to pectinase activity, it could be concluded that, in the fermentation for producing pectin, when the protopectinase activity is higher, Q is larger and the condition for producing pectin is better [23].
(1) Effect of fermentation time
The growth curve of strain CD-01, the effects of fermentation time on the protopectinase activity and pectinase activity are shown in Figs.5(a) and (b), which indicate that, except the vicinity of 36 and 56 h, the trend of the growth curve of strain CD-01 and the pectinase activity curve coincides in the exponential phase, which can be explained that the enzyme amount may change along with the growth of biomass (the amount ofAspergillus niger CD-01 in Czapek-Dox medium). Moreover, as shown in Fig.5(c), the change rate of pectin content in the medium at 36 and 56 h is much less than other points, which is caused by the catabolite inhibition. Therefore, the low activity of pectinase at 36 and 56 h should be caused by the catabolite inhibition [21]. On the other hand, the induction of the catabolite may cause the high activity of protopectinase at 36 and 60 h [22].
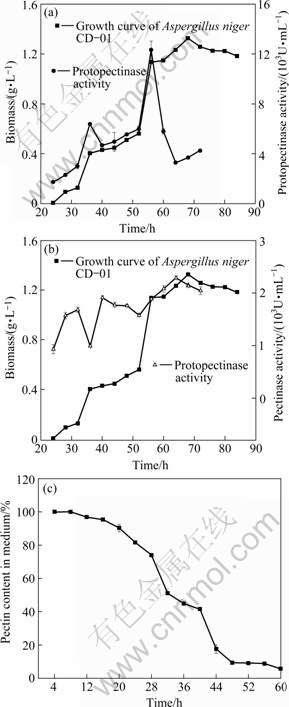
Fig.5 Growth curves of strain CD-01 and effects of fermentation time on activities of protopectinase (a) and pectinase (b), and change of pectin content in medium (c)
The effects of fermentation time on the pectin yield (data from pectin extraction experiment) and Q are shown in Fig.6, which indicates that there are two peaks at 36 and 56 h on the curve of Q, and the pectin yield is the highest at 36 h but decreases rapidly after that. Considering the fact that the strain, as shown in Fig.5(a), grew rapidly after 36 h, it can be derived that the decline of the pectin yield could be caused by the rapid consumption of pectin after 36 h. The two curves in Fig.6 are corresponding perfectly each other in the period of 28-44 h.
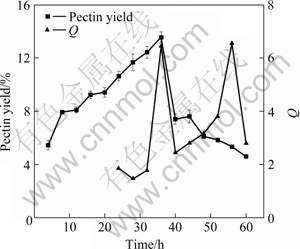
Fig.6 Effects of fermentation time on Q and pectin yield
In summary, the best time for sampling the sample could be 36 h when both Q and protopectinase activity are high, instead of 56 h, because at that time the activity of pectinase is high but pectin yield is low.
(2) Effect of fermentation temperature
The effects of fermentation temperature on Q and activities of protopectinase and pectinase are shown in Fig.7, indicating that the optimum temperatures for protopectinase and pectinase are 35 and 30 ℃, respectively, the values of Q are higher at 35, 40 and 45 ℃, and the protopectinase activities at 40 and 45 ℃ are much lower than that at 35 ℃. Therefore, it can be derived that the optimal fermentation temperature is 35 ℃, while temperature at 40 and 45 ℃ are not suitable for producing pectin by fermentation.
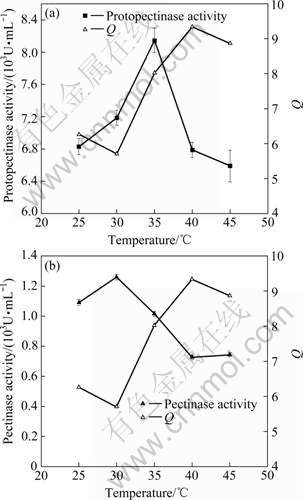
Fig.7 Effects of fermentation temperature on Q and activities of protopectinase (a) and pectinase (b)
(3) Effect of pH value
The effects of pH values on Q and activities of protopectinase and pectinase are shown in Fig.8. It can be seen that the activities of protopectinase and pectinase are the highest at pH 3 and pH 4, respectively, and the value of Q is higher at pH 4 than that at pH 3, meaning that pH 4 is better for producing pectin. Though the values of Q are much higher at pH 6 and 7 than that at pH 4, the activity of protopectinase, however, is much lower than that at pH 4. So, the best pH value for this test is pH 4.

Fig.8 Effects of pH value on Q and activities of protopectinase (a) and pectinase (b)
(4) Effect of nitrogen source
Four kinds of inorganic nitrogen source including NH4+ or NO3- and one kind of organic nitrogen source urea were chosen for the effect of nitrogen sources investigation. The results are shown in Fig.9, indicating that urea and NH4+ are better than NO3- for the protopectinase in fermentation, and the nitrogen source urea gives the highest protopectinase activity and the values of Q. It can be derived that the optimal nitrogen source is urea.
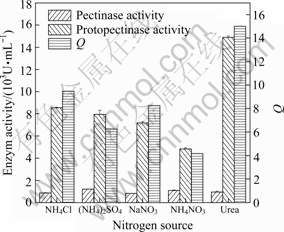
Fig.9 Effects of different nitrogen sources on Q and activities of protopectinase and pectinase
3.3 Orthogonal test
In order to get the interaction of the above factors on Q and the activities of protopectinase and pectinase, the orthogonal test in 3 levels, L9(34) was chosen for further optimization.
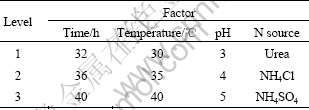
The levels of these factors are listed in Table 1. The range analysis in the orthogonal experiments are shown in Table 2, in which 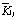 ,
,  , and
, and 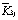 are the average of the responses corresponding to levels 1, 2, and 3, respectively, and R=max(
are the average of the responses corresponding to levels 1, 2, and 3, respectively, and R=max( )-min(
)-min( ). The larger the value of
). The larger the value of  , the better the condition, and the larger the value of R, the greater effect the factor. Based on this idea, according to the values of
, the better the condition, and the larger the value of R, the greater effect the factor. Based on this idea, according to the values of  in Table 2, the best parameter conditions for the highest protopectinase activity and Q are as follows: time 36 h, temperature 35 ℃, pH 5, and urea as nitrogen source; while the best parameter conditions for pectinase activity are as follows: time 36 h, temperature 30 ℃, pH 5 and NH4Cl as the nitrogen source.
in Table 2, the best parameter conditions for the highest protopectinase activity and Q are as follows: time 36 h, temperature 35 ℃, pH 5, and urea as nitrogen source; while the best parameter conditions for pectinase activity are as follows: time 36 h, temperature 30 ℃, pH 5 and NH4Cl as the nitrogen source.
Table 1 Levels and their values for factors of orthogonal experiments
On the other hand, according to the values of R in Table 2, it can be derived that the influence of the factors on pectinase activity is RpH>Rtemperature>Rtime> RN source, meaning that the major effect also comes from pH. The effect of the factors on protopectinase activity is Rtemperature>Rtime>RpH>RN source, meaning the major effect comes from temperature. The effect of the factors on Q is Rtemperature>Rtime>RN source>RpH, meaning the major effect also comes from temperature.
Table 2 Range analyses of activities of protopectinase and pectinase in orthogonal experiments

In variance analysis, F0.10=9.00 is taken as a reference value, and F>9.00 means a significant effect of that factor. According to the results of variance analysis in Table 3, factor temperature, whose F values for both protopctinase activity and Q are greater than 9.00, has a significant effect on protopectinase activity and Q, while factor pH has a significant effect on pectinase activity.
Table 3 Results of response variance analysis
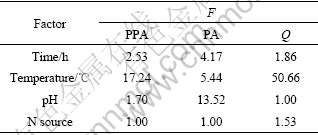
It can be concluded that the optimum condition for producing pectin is as follows: time 36 h, temperature 35 ℃, pH 5 and urea as nitrogen source, and the effect of factor temperature is significant.
Under the best condition, i.e., 36 h, 35 ℃, pH 5 and urea as N source, when 6.67 g dry citrus peel was added into 100 mL liquid medium, in which 10% of inocula with 1.75×106 cells/mL of spore concentration is inoculated, the pectin yield is 24.5% (on dry peel).
4 Conclusions
(1) A protopectinase-overproducing strain for pectin production was isolated from a pit soil dumped with perished orange in Changde City, Hunan Province, China. According to the morphology features and gene sequence analysis, the strain falls into the species Aspergillus niger. We name it Aspergillus niger CD-01.
(2) The fermentation experiment reveals that Aspergillus niger CD-01 is a protopectinase- overproducing strain and the optimal condition is as follows: time 36 h, temperature 35 ℃, pH 5 and urea as nitrogen source.
(3) The pectin yield under the optimal condition is 24.5%, as high as the yield of other physical and chemical methods until now. The strain Aspergillus niger CD-01 is suitable for pectin extraction. Besides the production of pection protopectinase and pectinase could also be obtained from the fermentation liquid, which is an advantage compared with traditional method.
References
[1] YANG Hong-shun, AN Hong-jie, LI Yun-jie. Manipulate and stretch single pectin molecules with modified molecular combing and fluid fixation techniques [J]. Eur Food Res Technol, 2006, 223(1): 78-82.
[2] TU Guo-yun, WANG Zheng-wu, WANG Zhong-ni. Preparation and application of pectin [J]. Food and Drug, 2007, 9(6): 50-55. (in Chinese)
[3] TAKUO S, MINORU O. Microbial production of pectin from citrus peel [J]. Applied and Environmental Microbiology, 1980, 39(4): 908-912.
[4] YOU Hua, LU Zhao-xin, FENG Hong-xia. Advance in micro- organism protopectinases research [J]. Industrial Microbiology, 2002, 32(1): 50-53. (in Chinese)
[5] ESQUIVEL J C C, HOURS R A, VOGET C E, MIGNONE C F. Aspergillus kawachii produces an acidic pectin releasing enzyme activity [J]. Journal of Bioscience and Bioengineering, 1999, 88(1): 48-52.
[6] LIU Hai-han, XU Gang, TIAN Qing-hua, GUO Xue-yi. Technical review on pectin extraction from the citrus pericarp[EB/OL]. [2006-05-29]. http://www.paper.edu.cn/paper.php?serial_number= 200605-361. (in Chinese)
[7] LIU Jia-jia, ZHAO Guo-ling, WANG Hui, ZHANG Xiao-hua. Extraction process of chlorogenic acid in flos lonicerae by enzymatic treatment [J]. Journal of Central South University of Technology, 2002, 9(4): 246-249.
[8] SAKAMOTO T, HOURS R A, TAKUO S. Purification, characterization, and production of two pectic transeliminases with protopectinase activity from Bacillus subtilis [J]. Bioscience, Biotechnology, and Biochemistry, 1994, 158(2): 353-358.
[9] LIU Chang-xiong, CHEN Shi-hua, WU Xing-quan. Research on the DNA extraction method from Trichoderma viride [J]. Journal of Henan University of Technology: Natural Science Edition, 2005, 26(6): 33-36. (in Chinese)
[10] ROSA L F, MTERESA F E, AMPARO Q. Identification of species of the genus candida by analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers [J]. Antonie Van Leeuwenhoek, 2004, 85(3): 175-185.
[11] ZHOU Hong-bo, ZEN Xiao-xi, LIU Fei-fei, QIU Guan-zhou, HU Yue-hua. Screening, identification and desilication of a silicate bacterium [J]. Journal of Central South University of Technology, 2006, 13(4): 337-341.
[12] CHEN Y C, EISNER J D, KATTAR M M, RASSOULIAN- BARRETT S L, LAFE K, YARFITZ S L, LIMAYE A P, COOKSON B T. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes [J]. Journal of Clinical Microbiology, 2000, 38(6): 2302-2310.
[13] YOU Hua. Screening of strain producing protopectinase, optimization of fermentation conditions and characteristics of protopectinase [D]. Nanjing: Nanjing Agricultural University, 2002. (in Chinese)
[14] MA Dong-ping, LIU Wei-juan, WEI Qing, LIU Gang, YANG Ting-shu. Determination of pectin in tobacco with 3,5-dinitrosalicylic acid colorimetry [J]. Tobacco Science & Technology, 2006, 28(8): 38-41. (in Chinese)
[15] YOU Xin-xia, QIU Nong-xue. Study on pectin content in apple pomace by carbazole spectrophotometric determination method [J]. Sichuan Food and Fermentation, 2007, 43(1): 19-22. (in Chinese)
[16] WANG Xiao-min, WU Wen-long, LU Lian-fei, LI Wei-lin, QU Le-wen. Study on the spectrophotometric analysis of pectinase activity [J]. Science and Technology of Food Industry, 2007, 28(5): 227-229. (in Chinese)
[17] ZOU Qi-meng. Decolorization of anthraquinone dyes by a biosorptive strain GX2 [D]. Tianjing: Nankai University, 2000.
[18] SHUTTON D, FOTHERQILL A, RINALDI M. Guide to clinically significant fungi [M]. Baltimore: Williams and Wilkins, 1998.
[19] PRESCOTT L, HARLEY J, KLEIN D. Microbiology [M]. New York: McGraw-Hill, 2002.
[20] XIA Jin-lan, PENG An-an, HE Huan, YANG Yu, LIU Xue-duan, QIU Guan-zhou. A new strain Acidithiobacillus albertensis BY-05 for bioleaching of metal sulfides ores [J]. Trans Nonferrous Met Soc China, 2007, 17(1): 168-175.
[21] DIAZ-GODINEZ G, SORIANO-SANTOS J, AUGUR C, VINIEGRA-GONZALEZ G. Exopectinases produced by Aspergillus niger in solid-state and submerged fermentation: a comparative study [J]. Journal of Industrial Microbiology and Biotechnology, 2001, 26(5): 271-275.
[22] YOU Hua, LU Zhao-xin, FENG Hong-xia. Study on the optimum fermentation condition for production of protopectinase by Aspergillus sp. [J]. Microbiology, 2003, 30(1): 26-30.
[23] CAVALITTO S F, HOURS R A, MIGNONE C F. Quantification of protopectinase SE, an endopolygalacturonase with pectin-releasing activity from Geotrichum Glebahnii [J]. Biotechnology Techniques, 1999, 13(6): 385-390.
___________________
Foundation item: Projects(50621063, 50674101) supported by the National Natural Science Foundation of China
Received date: 2008-02-28; Accepted date: 2008-06-12
Corresponding author: XIA Jin-lan; Professor, PhD; Tel: +86-731-8836944; E-mail: jlxia@mail.csu.edu.cn
(Edited by YANG You-ping)